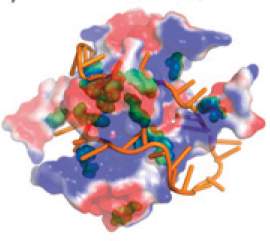
As the helix opens, various proteins slide along the double-stranded DNA searching for their unique binding spot, and the “back brace” proteins, too, slide on the single-stranded DNA. Usually interactions between sliding proteins and DNA are based on differences in electrical charges: positive for the proteins and negative for the DNA. The new Weizmann Institute model has revealed that in the case of the “back brace” proteins, the situation is more complex. These proteins also interact with the single DNA strand chemically, via amino acids possessing structures called aromatic groups. Through these two types of interaction, the “back brace” proteins and the single strand wrap around each other until they form a stable complex.
With the help of their model, the Weizmann researchers correctly predicted the precise configuration of such complexes: Their predictions were validated by comparison with actual structures that had earlier been solved experimentally for crystallized protein-DNA complexes.
The theoretical backing offered by the Weizmann study can facilitate new experimental investigations. In particular, says Levy, scientists can now investigate whether previous discoveries made about interactions between proteins and double-helix DNA also apply to protein interactions with single DNA strands. For example, in the past few years he and his team had helped to resolve the speed-stability paradox: how proteins sliding along the double helix manage to swiftly find appropriate spots for binding, yet at the same time to create bonds that are sufficiently strong to support molecular interactions. Using computational analysis, the team had identified geometrical and energetic parameters that enable the proteins to bind with the DNA at the optimal speed and strength.
By shedding new light on how the DNA functions at the molecular level, this study advances the understanding of biological processes that form the very basis of life and may lead to improved biotechnological applications based on protein-DNA interactions.