Are you a journalist? Please sign up here for our press releases
Subscribe to our monthly newsletter:
Genes may contain the blueprint for life, but they’re written more like coded messages in a spy thriller: Short segments of DNA carrying instructions for protein formation are imbedded in a longer text, and interspersed with “filler” DNA that has no known function. The picture becomes still more complicated when one realizes that the useful “characters” in the genetic code may be pieced together, or spliced, in different combinations. “Alternative splicing” allows relatively few genes to give rise to a great number of protein structures.
Since the discovery of RNA splicing, around 25 years ago, scientists have worked to understand how the right sequences are lifted out and strung together to make a coherent set of instructions. Both straightforward and alternative splicing take place in the “spliceosome,” situated in the cell nucleus. A large complex of proteins and short strands of RNA, the spliceosome distinguishes the beginnings and ends of coded segments, precisely cutting and “stitching” them together.
A husband-and-wife team, Prof. Ruth Sperling of the Hebrew University of Jerusalem’s Genetics Department and Prof. Joseph Sperling of the Weizmann Institute’s Organic Chemistry Department, has produced the most detailed 3-D representation of the spliceosome’s structure to date. Rather than follow others’ attempts to observe spliceosomes created in test tubes, the Sperlings and team members Maia Azubel, Ruth’s graduate student, and Sharon Wolf of the Institute’s Chemical Research Support Department managed to take spliceosomes directly from living cells and examine them under an electron microscope.
The living spliceosome presented them with a challenge. In cells they come packaged in sets of four identical modules strung together like beads on a strand of RNA, each a miniature spliceosome capable of splicing on its own. The connections between the modules tend to be flexible, allowing the position of the units to vary in relation to one another, and making pinning down a definitive shape and structure for the whole complex close to impossible.
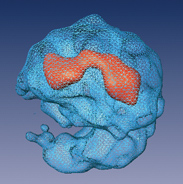
The team found a way to cut the RNA connections between the modules without harming the integral short strands of RNA that are essential to the splicing process, so they could study them individually. Split-second freezing at very low temperatures allowed the scientists to view the spliceosome units in a state as close to natural as possible. From thousands of images, each at a slightly different angle, a composite 3-D structure of the spliceosome was built up.
The revealed structure has two distinct, unequal halves surrounding a tunnel. The larger part appears to contain proteins and the short segments of RNA, while the smaller half is made up solely of proteins. On one side the tunnel opens up into a cavity, which the researchers think functions as a holding space for fragile RNA waiting to be processed in the tunnel itself.
What they didn’t see may be as important as what they saw. Whereas researchers examining splicing in test tubes saw evidence of a complicated sequence of events in which the spliceosome machinery assembles itself anew for each splicing job, the team’s investigations of spliceosomes from live cells found splicing to take place in preformed machines. This fits in with what is known about the way cells optimize their workload. “It’s much more efficient to have a machine on hand, ready to go, than to build a new one each time,” they noted.
Prof. Joseph Sperling’s research is supported by the Helen and Milton A. Kimmelman Center for Biomolecular Structure and Assembly; the J & R Center for Scientific Research; the Joseph and Ceil Mazer Center for Structural Biology; and Lois Zoller, Chicago, IL. Prof. Sperling is the incumbent of the Hilda Pomeraniec Memorial Chair of Organic Chemistry.