Are you a journalist? Please sign up here for our press releases
Subscribe to our monthly newsletter:
Cells can get lost in a crowd too, making it difficult for scientists to detect and study them. In the tissues that line the inner surfaces of organs such as the small intestine, liver or kidneys, certain cell types with unique attributes are grossly outnumbered by their more “common” neighbors. Yet despite their small numbers, these rare cells play a significant role in maintaining our health. In the lining of the small intestine, for example, goblet cells, aptly named after their shape, make up only 10 percent of the tissue but release all the mucus needed to protect it. And enteroendocrine cells, which account for less than 1 percent of the intestinal lining, secrete hormones that are required for proper metabolism, such as those involved in sugar regulation.
To study such rare cell types, Prof. Shalev Itzkovitz and his team in the Molecular Cell Biology Department at the Weizmann Institute of Science developed an approach called “Clump Sequencing,” or “ClumpSeq” for short. It allows researchers to track the behavior of rare cell types in a variety of different tissues, constructing elaborate cell atlases as they go.

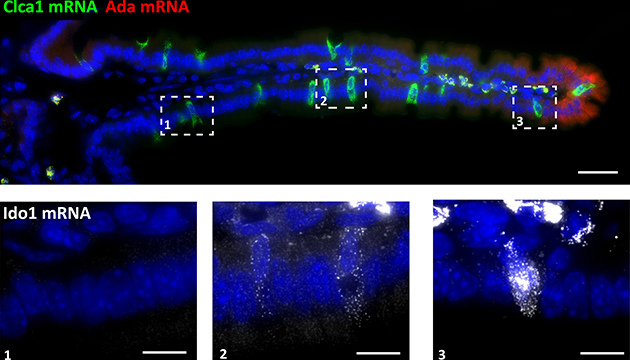
The new approach relies on abundant cell types – for example, the nutrient-absorbing enterocytes, which compose close to 90 percent of the intestinal lining – to track down rare cells, such as the goblet or enteroendocrine cells, which are difficult to find and characterize using existing methods. Over the course of their short lives, enterocytes move through the inner lining of the small intestine, a folded structure made of repeating crypts and tiny finger-like protrusions facing the intestinal cavity called villi (singular: villus). Previous work from Itzkovitz’s group has shown that as the enterocytes migrate from the deep stem-cell-harboring crypt and along the villus before being shed off at its tip (a process that takes approximately four days to complete), they constantly change their behavior. This means that at different locations they express different genes, as can be deduced from the types of messenger RNA molecules, or mRNAs, these genes make. And this in turn means that at each location it is possible to learn a great deal about the function of enterocytes by observing their mRNA.
""ClumpSeq" is a method developed by Itzkovitz and his team for analyzing rare cell types that relies on defining genetic landmarks similar to the use of geographical landmarks when navigating
But when the scientists tried to characterize rare cell types by examining their mRNA profiles, they ran into a problem. “When we would apply traditional analysis methods to analyze mRNA profiles in different tissue zones, the mRNA content of these rare cells would simply become diluted, owing to its small amount compared to that of enterocytes,” Itzkovitz says.

The researchers solved the problem by cutting the tissue into clumps of two to ten cells – hence the method’s name – and then analyzing the mRNA content of this small clump versus that of a larger chunk of tissue, knowing that one out of ten would likely be a rare cell type. They then scanned this RNA pool for “landmark genes” known to be expressed by the abundant enterocytes. Landmark genes are similar to geographical landmarks that we use when navigating, only instead of monuments or lampposts, they have unique mRNA signatures that are each associated with a particular zone in the tissue. By identifying these landmark genes, researchers were able to determine the zone from which each analyzed clump originated – either the crypt or any segment of the villus, from the bottom to its tip. Once they solved the entire clump’s location, the researchers started scanning for and identifying the landmark genes of the rare cells and picked up on their unique gene expression patterns.
Applying this approach to the analysis of the mouse intestine, the researchers successfully demonstrated its potential for characterizing rare cell types. They discovered, for example, that goblet cells changed their behavior almost as much as enterocytes do: About a third of their genes altered their expression as the “goblets” were migrating up the villus. In particular, genes that regulate the activity of the immune system were often expressed by goblet cells exclusively at its tip. “Since the top region of the villus is full of bacteria and also the point where cells shed off, leaving a microscopic opening in the tissue,” Itzkovitz explains, “it makes a lot of sense for cells located in this area to have some regulation over the immune system, to prevent inflammation from forming where it shouldn’t.” They also found that some types of enteroendocrine cells that were previously thought to be migrating continuously along the villus, similar to enterocytes, actually only appear at the bases of the villi.
ClumpSeq can be applied to studying the roles played by various rare cell types in the healthy organism and how their function goes awry in perhaps not so rare diseases. For example, tuft cells are rare lining cells that modulate the immune response in tissues such as the lungs and the gut. Characterizing their location-based behavior could help us to better understand disorders such as allergies, asthma and inflammatory bowel diseases. This approach can also be used to study cancerous tumor cells and how their behavior changes depending on whether they are near or far from the cells that compose the blood vessels providing oxygen and nutrients to the tumor.
The research team included postdoctoral fellows Dr. Rita Manco and Dr. Inna Averbukh, and Staff Scientist Dr. Keren Bahar Halpern – members of Prof. Itzkovitz’s group, in addition to Dr. Ziv Porat from the Life Sciences Core Facilities Department and Prof. Ido Amit from the Immunology Department.
Enteroendocrine cells are only 1% of the cells that line our gut, nevertheless, given the gut's extensive area it is the largest endocrine organ in our body.
Prof. Shalev Itzkovitz's research is supported by the Helen and Martin Kimmel Institute for Stem Cell Research; the Wolfson Family Charitable Trust; and the Dr. Beth Rom-Rymer Stem Cell Research Fund.