Are you a journalist? Please sign up here for our press releases
Subscribe to our monthly newsletter:
Every protein in our body has a unique vital function, while its malfunction can cause disease. That is why researchers want to study different aspects of their protein of choice: What is its function in the cell, organ or body? How does it interact with other proteins? How does its structure dictate its character?
For scientists seeking answers to these questions, there is the Dana and Yossie Hollander Center for Structural Proteomics in the Weizmann Institute of Science. Researchers from academia and industry from Israel and abroad – all utilize the Center’s specialized pipeline. Within the Center staff scientists, along with their expert technicians, engineer, express, purify, and supply clean proteins, and solve their 3-D crystal structure. The staff scientists find themselves involved in a variety of interesting projects together with research groups through all stages of a study, from the cloning of genes to understanding how the 3-D structure of a protein ties in to its function.
The Manipulators

Producing a protein that can be isolated in the lab usually requires some manipulations of its DNA sequences. Staff Scientists Drs. Tamar Unger and Yoav Peleg developed an innovative approach for gene cloning and mutagenesis that speeds up the process and is more efficient. The method is now used by scientists around the world for many applications in manipulating DNA. “I can now take any piece of DNA of choice and clone or make changes to it,” says Unger.“ The applications we developed significantly reduce the cost of making changes to DNA sequences and they do not require advanced equipment,” says Peleg.
After the changes have been applied to the gene of interest, the research generally requires large quantities of the protein encoded by the gene. Unger and Peleg and their team – Ada Dantes, Netta Segal-Gilboa and Meital Yona – are experts in the expression of these proteins in bacteria and yeast, as well as insect and human cells.
The Selectors

The engineered protein is expressed along with thousands of proteins in the cell. Thus the desired protein must be isolated and purified – a job that can best be defined as finding the needle in a haystack. So scientists turn to the expertise of Dr. Shira Albeck who, with the help of lab technician Jossef Jacobovitch, purify proteins using a range of techniques, taking advantage of a particular biochemical or biophysical feature of the protein, or its affinity for other proteins or ligands. The idea is to end up with a protein that is not only pure, but is stable and functional. That means the process must be conducted quickly, but with delicacy. The protein must always be satisfied with its conditions – otherwise its structure could be impaired and it might refuse to engage in the activities that are the subject of the research. “Characterization and stabilization are the name of the game in protein research,” says Albeck.
The Cracker

Staff Scientist Dr. Orly Dym knows what it means when her lab tech Shelly Rogotner yells her name in the Proteomics Center’s corridors: “It’s a sign that we have a protein crystal,” she says. Purified proteins come to her in solution; to reveal their structure in detail, it is necessary to turn them into crystals. Obtaining protein crystals is a long, tedious trial and error process by which different chemicals are added to the protein solution, until the team finds one that chases out the water and leaves the protein molecules "lined up like soldiers.” The crystal is then bombarded with x-rays; the scattering of the x-rays reveals the 3-D structure of the protein. That structure, together with its biochemical or biophysical features, can tell scientists how the protein functions, as well as how malfunctions cause disease.
The Investigations
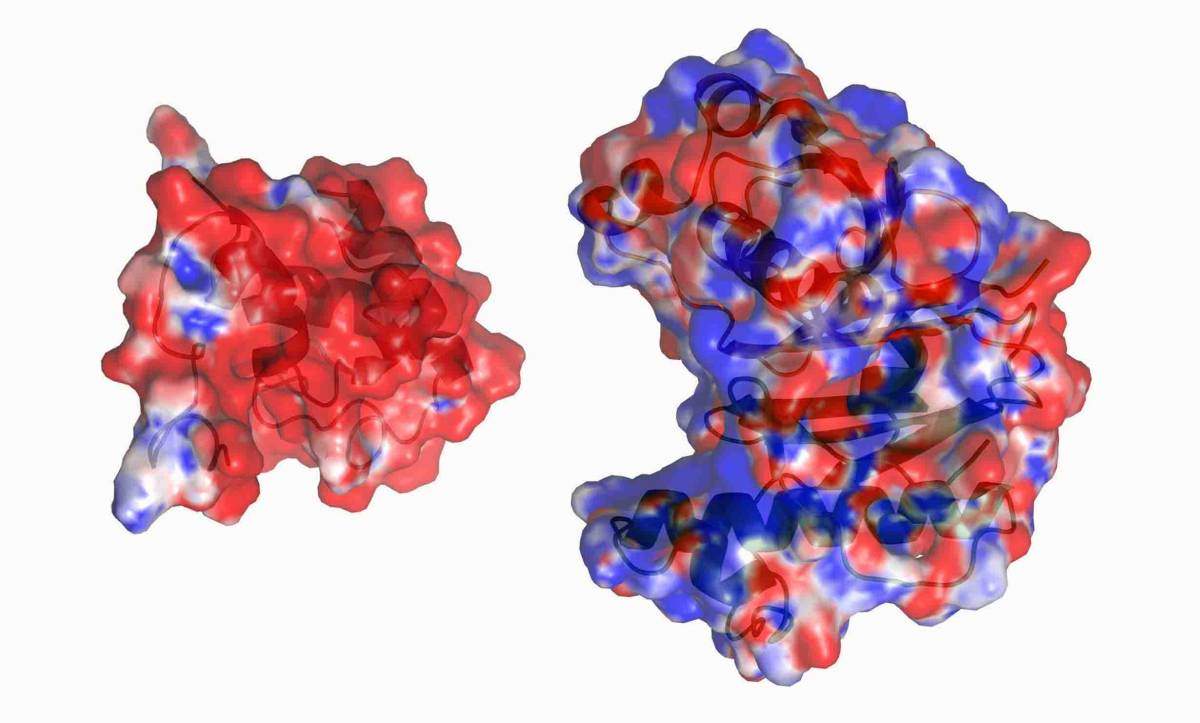
Over 100 proteins pass through the Dana and Yossie Hollander Center for Structural Proteomics each year. Over the years, the center has been involved in many fruitful collaborations with scientific groups at the Weizmann Institute. Dr. Sarel Fleishman of the Biomolecular Sciences Department designed several proteins with improved stability or activity. The Center’s staff scientists produced and determined the 3-D structure of these novel proteins to validate Sarel’s algorithm. Prof. Ed Bayer of the same department came to them in order to manipulate various components of the cellulosome – a protein complex for breaking down cellulose – to see how the organization of its various components affects the function of the whole complex.
The Center’s team worked with Prof. Zippora Shakked of the Structural Biology Department and Dr. Oren Zimhony of Kaplan Medical Center to understand the mechanism employed by an enzyme found in the tuberculosis bacterium to produce fatty acids. The bacterium needs these fatty acids to build its protective outer membrane. The researchers hope their findings may lead to new means of attacking this bacterium, which is currently resistant to many antibiotics.