Are you a journalist? Please sign up here for our press releases
Subscribe to our monthly newsletter:

Talk about embracing hardship: A single-celled archaea thrives amidst superheated gases spewed from openings in Earth’s crust. Researchers at the Weizmann Institute of Science, in collaboration with scientists from the United States, have discovered a mechanism used by this organism to survive inside steaming cracks in the ground on a Japanese island. The mechanism – a kind of heat-resistant gear for RNA – constitutes a previously unknown way by which cells can adjust to a changing environment. This discovery opens new directions for studying human health and disease.
For Thermococcus kodakarensis (a species of archaea), a scalding 50oC is uncomfortably cold. This microscopic organism, isolated from a solfatara – an opening in the ground that emits sulfurous gases – near the shore on Japan’s Kodakara Island, thrives at temperatures approaching the boiling point (100oC).
The organism attracted the attention of Weizmann Institute of Science researchers who were studying an ancient modification in the composition of RNA – one that has been preserved in the course of evolution. This modification is a chemical tag of a type that can affect the stability and structure of RNA. It is referred to as ac4C – that is, the attachment of an acetyl tag to the fourth position on an RNA component known as cytidine. Chemical tags on RNA molecules have diverse functions, altering the cell’s genetic code as well as RNA-based cellular machines.

Dr. Aldema Sas-Chen and colleagues in the laboratory of Dr. Schraga Schwartz of the Molecular Genetics Department, along with Dr. Jordan L. Meier of the National Cancer Institute in the United States, developed a method for chemically manipulating the RNA to make the ac4C “visible” when sequencing is performed. The scientists hypothesized that ac4C strengthens RNA molecules, and they started looking for cases in which extra strengthening could be advantageous. This brought them to Thermococcus kodakarensis, an organism whose RNA evidently requires a fair amount of strengthening, as this is a molecule that generally disintegrates in heat.
When the scientists applied their method to the RNA of this organism, they found astounding amounts of ac4C: Whereas human cells and yeast contain only a handful of these modifications, each heat-resistant, single-celled creature was found to have around 400 of them. In particular, a great many ac4C modifications were seen in the ribosome, the protein-making machinery that is built around folded RNA molecules. These molecules were shown to contain as many as 170 ac4C modifications. The scientists genetically altered Thermococcus kodakarensis to delete the enzyme that produces ac4C and found that the mutant organisms were vulnerable to heat, which suggested that ac4C is indeed essential for their survival at extremely high temperatures.
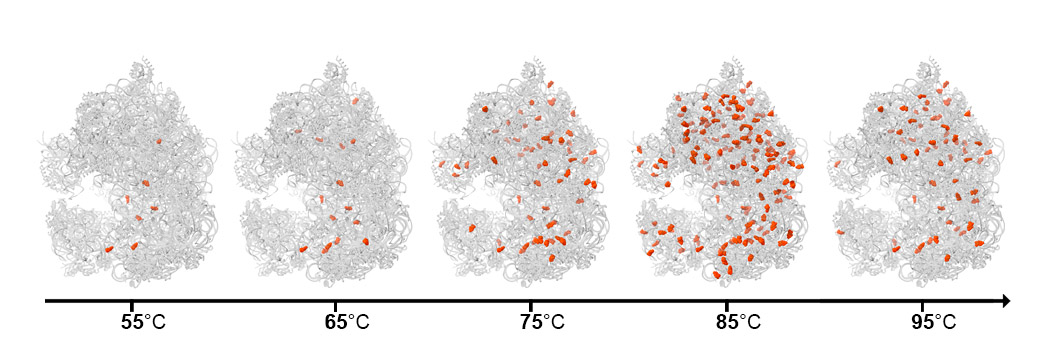
The addition of more and more ac4C is a dynamic adaptation mechanism – analogous to putting on layers of heat-resistant armor
Next, the scientists teamed up with the group of Dr. Moran Shalev-Benami of Weizmann’s Structural Biology Department to map out ac4C in greater detail and in three dimensions. Dr. Donna Matzov, from Shalev-Benami’s lab, used cryogenic electron microscopy to solve the atomic structure of ribosomes and to establish exactly where ac4C appears and what happens to the ribosome structure in its absence. She saw that in contrast to the human ribosome, in which the tag is only attached to two locations at the active site, in Thermococcus kodakarensis, it could be attached all over the ribosome. In addition, she found that the number of ac4C tags in the single-celled creatures increased as the temperature rose: There were hardly any at 50oC, but their number soared at around 80oC and continued to increase with more heating. This progressive attachment led the researchers to conclude that the addition of more and more ac4C is a dynamic adaptation mechanism – analogous to putting on layers of heat-resistant armor – to shield the ribosome from rising temperatures.
The study’s findings suggest that the ac4C modification constitutes a previously unknown cellular technique for adjusting to a changing environment. The method developed for measuring and mapping out ac4C may be employed in future studies to explore the modification’s potential role in a variety of changing contexts. This method may, for example, make it possible to check whether ac4C helps protect the RNA in human cells when we run a fever, or under other stressful conditions, such as those present in cancer and neurodegenerative diseases. In addition, the method may help in studies aimed at revealing whether mutations in the enzyme responsible for creating ac4C, which are found in at least two types of cancer and in a certain form of premature aging, contribute to these diseases.
Study participants included Ronit Nir, Ran Shachar and Jesse Hartmann from Weizmann’s Molecular Genetics Department; Justin M. Thomas, Kellie D. Nance, Keri M. Bryson, Chloe A. Briney and Supuni Thalalla Gamage from the National Cancer Institute in the United States; Masato Taoka, Yuko Nobe and Prof. Toshiaki Isobe from Tokyo Metropolitan University; Geraldy L.S. Liman, Brett W. Burkhart and Prof. Thomas J. Santangelo from Colorado State University; Michaella J. Levy, Michael P. Washburn and Laurence Florens from Stowers Institute for Medical Research, Kansas City, Kansas, USA; Ryan T. Fuchs and G. Brett Robb from New England Biolabs, Inc., Ipswich, Massachusetts, USA; Sunny Sharma from Rutgers University; and Qishan Lin from the University at Albany.
Dr. Schraga Schwartz's research is supported by the Ilse Katz Institute for Material Sciences and Magnetic Resonance Research; and the estate of Emile Mimran. Dr. Schwartz is the incumbent of the Robert Edward and Roselyn Rich Manson Career Development Chair in Perpetuity.
Dr. Moran Shalev-Benami's research is supported by the Ilse Katz Institute for Material Sciences and Magnetic Resonance Research; the Helen and Milton A. Kimmelman Center for Biomolecular Structure and Assembly; the Zuckerman STEM Leadership Program; the Joseph and Wolf Lebovic Lab; the Dov and Ziva Rabinovich Endowed Fund for Structural Biology; the Harmstieg New Scientist Fund; the Pearl Welinsky Merlo Foundation; Paul and Tina Gardner; and Dr. and Mrs. Donald Rivin.