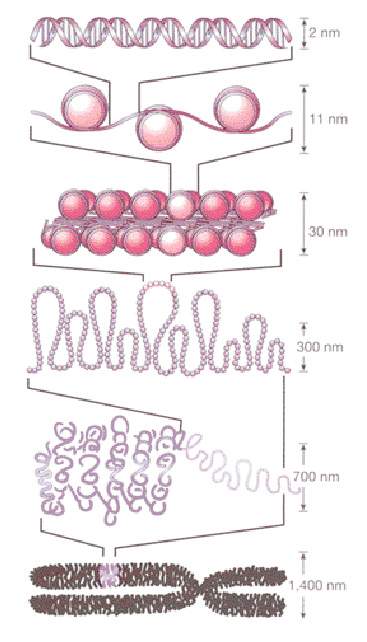
The most familiar image of DNA, the basic material of heredity, is that of two long strands of genetic “letters” strung together and twisted around each other in the famous double helix. But inside the cell nucleus, this double strand, which reaches over a meter in length when stretched out, is compressed into neat parcels. This DNA packaging is more than just a way of maintaining tidiness in the cell nucleus – it keeps much of our DNA locked away, preventing it from being easily copied.
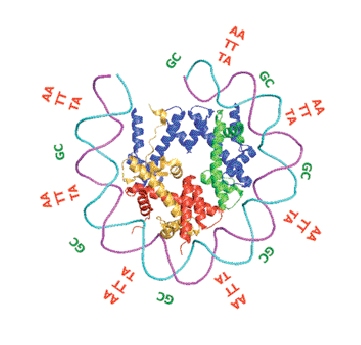
The DNA strand is first packaged into “stackable” units called nucleosomes, each about 150 base pairs in length (base pairs being the “letters” that make up the genetic sequence), which are compressed into tiny spheres around proteins. The nucleosomes are strung, bead-like, along the entire chromosome, separated by free areas of about 20 base pairs. The precise location of the nucleosomes along the double strand plays an important role in the cell’s day-to-day function: Access to nucleosome-wrapped DNA is blocked for many proteins, including those responsible for some of life’s most basic processes. Among these excluded proteins are factors that initiate DNA replication, the transfer of genetic information from DNA to RNA and the repair of damaged DNA. In other words, the positioning of nucleosomes limits the genetic segments in which these processes can take place to the brief nucleosome-free areas.
What determines how and where a nucleosome will be positioned along the DNA sequence? Scientists have disagreed for years whether the placement of nucleosomes in live cells is controlled by the genetic sequence itself. In an article published recently in Nature, Dr. Eran Segal and research student Yair Field of the Computer Science and Applied Mathematics Department of the Weizmann Institute proved that the DNA sequence indeed encodes “zoning” – that is, information on where to place nucleosomes. They managed, together with colleagues from Northwestern University in Illinois, to crack the genetic code that sets the rules for where on the DNA strand the nucleosomes will be situated. After they successfully characterized this code, they were able to accurately predict a large number of nucleosome positions in yeast cells, purely on the basis of the DNA sequence.
Segal and his colleagues accomplished this by examining around 200 different nucleosome sites on the DNA and asking whether their sequences had anything in common. Mathematical analysis revealed similarities between the nucleosome-bound sequences and eventually uncovered a specific “code word.” This “code word” consists of a periodic signal that appears every 10 bases on the sequence. The regular repetition of this signal helps the DNA segment to bend sharply into the spherical shape required to form a nucleosome. To identify this nucleosome positioning code, the research team used probabilistic models to characterize the sequences bound by nucleosomes; they then developed a computer algorithm to predict the organization of nucleosomes along an entire chromosome.
The team’s findings provided insight into another mystery that has long puzzled molecular bio-logists: How do cells direct the proteins that regulate genetic processes to their intended sites on the DNA, rather than to the many similar, but functionally irrelevant sites along the genomic sequence? The short binding sites do not themselves contain enough information for these proteins to discern among them. The scientists showed that basic information on the functional relevance of a binding site is at least partially written into the nucleosome code: The intended sites are found in nucleosome-free segments, thereby allowing them to be accessed by the proteins. In contrast, spurious binding sites with identical structures that could potentially sidetrack these proteins are conveniently situated in segments that form nucleosomes, and are thus mostly inaccessible.
Since the packaging proteins that form the core of the nucleosome are among the most highly conserved throughout evolution, the scientists believe that the genetic code they identified should be found in many organisms, including humans. Several diseases, among them cancer, are typically accompanied or caused by mutations in the DNA, and such mutational processes may be influenced by the relative accessibility of the DNA to various proteins and by the organization of the DNA in the cell nucleus. The scientists believe, therefore, that the nucleosome positioning code they discovered may significantly aid researchers in their attempts to understand the mechanisms underlying many diseases.
For Segal, the fact that computational modeling methods were crucial to these findings may have major implications for further research: “Often, a model yields insights that, in hindsight, could have been obtained by simple statistical analysis. In this work, our modeling approach played a key role in the discovery. It was only after we devised and applied our algorithm that we were able to obtain the biological insights that led to making successful predictions and proving that genomes do indeed encode, at least in part, nucleosome positions.”
Dr. Eran Segal’s research is supported by the Willner Family Leadership Institute for the Weizmann Institute of Science; the Arie and Ida Crown Memorial Charitable Fund; and the Estelle Funk Foundation.
 Yair Field and Dr. Eran Segal. Location%2C location%2C location.jpg)

