Are you a journalist? Please sign up here for our press releases
Subscribe to our monthly newsletter:

The smallest nose yet could consist of a single molecule. Prof. David (Didi) Margulies and his group at the Weizmann Institute of Science are demonstrating nano-sized artificial noses that work something like real noses − that is, they can sense a number of different molecules in the environment and combine this information into a “scent.” What’s more, these molecular noses can “smell” inside a living cell.
Margulies, who is in the Institute’s Organic Chemistry Department, explains that the artificial, chemical-based noses that exist today generally contain arrays of electronic sensors, each of which is coated in a different material that can bind a different class of molecule. Assimilating the readings from the different sensors, as our brains do with the signals from the receptors in our noses, is what produces the scent. However, these types of devices are too large for use in many fields, for example cell biology, in which detection takes place on the microscopic level.
In research that was recently published in Nature Nanotechnology, Margulies and his group created a nanodevice that combines an array of sensors on a single molecule. This tiny device can bind to multiple molecules and report on them by glowing in different colors. “This could give us a new analytical tool to provide information that we cannot access today on the chemical composition of micro environments such as cells,” says Margulies.
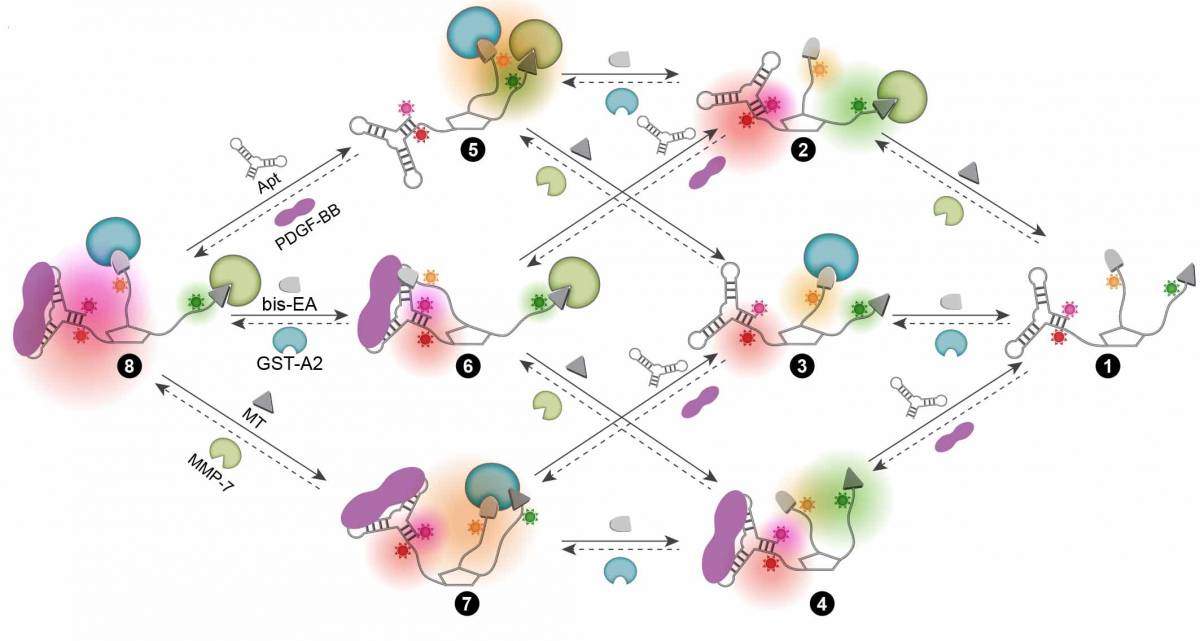
To create their nano-sized artificial nose, Margulies and his group designed a complex molecule on which several different molecular binders were linked to fluorescent groups that glow in different colors when binding takes place. Thus the molecular nose shines in different colors – a unique light signature – for every combination of substances it meets. This molecular device can be used again and again, and it can follow dynamic processes and change the light signature it emits accordingly.

In the present study, the molecular nose contained binders for three different protein families that have important functions in the cell and are implicated in disease. These, known by the abbreviations GST, MMP and PDGF, play a role in the development of cancer, among other things. Dr. Leila Motiei in Margulies’ group explains that the system needs to be at least partly specific (hence the three binders): “In a cell there are so many different proteins. If we did not adjust the system to bind specific protein groups, it would be like trying to identify individual odor molecules in a dumpster.”
The researchers first tried out their tiny nose by sampling these proteins in bodily fluids, for example, blood and urine. They then turned to their goal: Sniffing out the proteins’ activities inside single cells. “We caused cells in the lab to express certain proteins and inserted the molecular sensors in these. Indeed, we could identify the light signature for each of the different cells,” says research student Zohar Pode. “We then observed these cells as we changed their environments, creating inflammation-provoking situations or depriving them of oxygen. And here we observed new light signatures that reflected the changes in their states.”
What’s more, these molecular noses can “smell” inside a living cell
Margulies says that one of the possible uses of the molecular nose might be in drug development. This was demonstrated by the ability of the system to identify new inhibitors. In addition, devices such as the ones the team developed could be incorporated into medical tests for early disease discovery, and they may have many applications beyond the biomedical: “This opens up possibilities for sensing various other combinations of chemicals in the microscopic world,” says Margulies.
Also participating in this study were Ronny Peri-Naor, Joseph Georgeson, Dr. Tal Ilani of the Structural Biology Department, Vladimir Kiss of the Biomolecular Sciences Department, Dr. Tamar Unger of the Israel Structural Proteomics Center, and Dr. Barak Markus and Dr. Haim Barr of the Nancy and Stephen Grand Israel National Center for Personalized Medicine.
Prof. David Margulies's research is suppoorted by the Helen and Martin Kimmel Center for Molecular Design.