Are you a journalist? Please sign up here for our press releases
Subscribe to our monthly newsletter:
Chop it up, unzip it, make numerous copies of it in less than an hour, or even edit it, slipping in a trait for, say, bacterial resistance or insulin production. Seems like there's no end to the stuff one can do nowadays to DNA. And future aims are no less extraordinary, like the attempt to harness DNA as wiring for microelectronics.
All this, however, calls for a greater understanding of the physical properties of DNA, including the way in which it 'unzips' itself when exposed to heat a process called denaturing.
The shape of the DNA molecule has been known since 1953 when American microbiologist James Watson, then only 25, and English physicist Francis Crick discovered its double helical structure, earning them the 1962 Noble Prize. (They shared the prize with Maurice Wilkins of Kings College, London, who had solved the problem independently at the same time.)
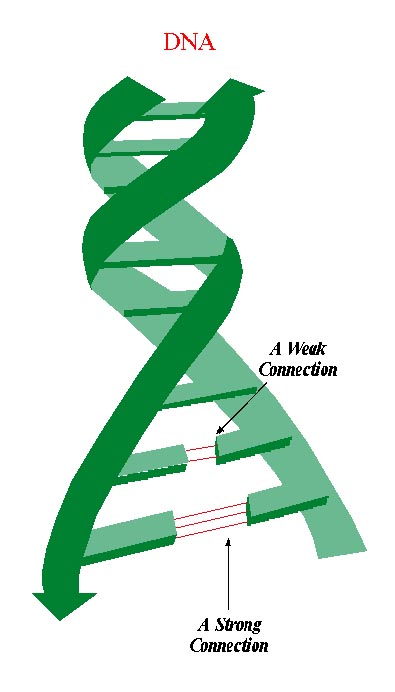
The key they discovered was chemistry. Shaped like a spiral staircase, the DNA molecule consists of two winding strands of alternating sugar and phosphate molecules. But the most interesting part of the molecule from a genetic perspective is not these strands but rather the way in which they are linked. Four nitrogen-containing bases: adenine, cytosine, guanine, and thymine (referred to as A, C, G, and T) wind up the helix, forming 'steps' as each base pairs up with a counterpart on the other strand. The sequence is far from random: A always pairs up with T, and C with G. Indeed, Watson and Crick realized that the strict chemical rules dictating the nature of these cross-links underlie the marvel of the DNA molecule. The specific sequence of A's, T's, C's, and G's along each DNA strand actually constitutes a code translated by cell machinery to synthesize proteins. Likewise due to these chemical rules, DNA is capable of duplicating itself with generally remarkable precision, unzipping the helix and using each strand as a template to direct the formation of a companion strand.
Later studies showed that the DNA molecule also 'unzips' itself (albeit with a drastically different outcome) when exposed to temperatures of about 70°C. During this separation process, loops form in certain parts of the genetic material but not in others. Why is this so? Scientists studying this physical phenomenon discovered that it stems from differences in the strength of the nucleotide bonds. The A-T bond is weaker than the C-G one. As a result, genetic sequences containing more A-T bonds will come apart faster and more easily than those rich in C-G bonds.
And this finding, made in the late fifties, years before advanced genetic sequencing methods were devised, stirred up a great deal of excitement. Scientists hoped to use statistical data on the location of these loops to backtrack to the DNA molecule itself, gaining information about its genetic sequence. This is where physicists studying different types of phase transitions became interested. The scientists, including the late Prof. Shneior Lifson of the Weizmann Institute, examined DNA models containing only A-T nucleotide bonds and showed that in these models, the transition from normal to 'loopy' DNA structure occurred gradually, with the loop size continuously increasing. However, experimental observations of loop formation in genetic material disproved this finding. The loops, they showed, did not increase in a slow and tidy fashion, since the DNA strands actually unzipped suddenly.
This frustrating contradiction in scientific knowledge persisted for more than 30 years, until graduate student Yariv Kafri of the Institute's Physics of Complex Systems Department, Prof. David Mukamel, Dean of the Physics Department, and visiting scientist Prof. Luca Peliti of the University of Napoli in Italy decided to take another look. They discovered that all the theoretical models used to describe the phenomenon contained a major flaw: they allowed adjacent DNA loops to overlap. Yet such overlapping never takes place in nature, since the DNA strands electrically repel each other.
This insight led the Institute scientists to study the possible ways in which DNA loops can arrange themselves in space while avoiding any overlapping between different loops and between the folded strands of the same loop. To describe the range of possible loop interactions, the researchers used mathematical knowledge gained from the study of various polymer networks. Once the avoidance of overlapping was taken into account with the help of these precise calculations, the modified theoretical models began to fit with experimental observations. The researchers predicted that when exposed to heat, transitions from a normal genetic sequence to loop formation happen suddenly just as in nature a phenomenon observed in laboratories around the world.
An accurate description of DNA may help in developing diverse applications, including a surprising partnership between microelectronics and natural molecules. The idea is to integrate organic molecules into microelectronics, to replace conventional components such as transistors, memory elements, and wires. This merger, say experts, may yield computer components thousands of times more compact than those presently available.
Prof. Mukamel holds the Harold J. and Marion F. Green Professorial Chair.