Are you a journalist? Please sign up here for our press releases
Subscribe to our monthly newsletter:

Have you ever tried to touch your nose with your finger when your eyes are closed? Most of us can do this with great precision. In research spanning decades, Senior Research Fellow Dr. Ditsa Levanon has been working in the lab of Prof. Yoram Groner of the Weizmann Institute of Science’s Molecular Genetics Department investigating the molecular developmental processes that grant us this ability. “Nearly everyone is familiar with our five senses – sight, sound, smell, touch and taste – but few are aware that we have what is sometimes referred to as a ‘sixth sense’ – the position sense,” says Groner. Such “body awareness,” otherwise known as proprioception, allows us to sense the position and movement of our limbs and our body.
Levanon and her colleagues in Groner’s lab found that a protein known as transcription factor Runx3 is an essential regulator of specialized sense neurons –TrkC neurons – which are involved in proprioception. “We found that when Runx3 activity is lost in mice, TrkC neurons die and the mice become ataxic: They are unable to place their legs in the correct position and lose the ability to control their movements and to balance,” says Levanon.
In the peripheral nervous system the Runx3 transcription factor turns out to be specific for TrkC neurons, but not for such closely related neurons as those for pain or touch. Levanon: “The question we are now trying to answer is: What exactly switches the Runx3 gene ‘on’ and ‘off’ at the right place and time, and how does this lead to the specific formation of TrkC neurons?”
Levanon and colleagues Drs. Elena Apple, Kira Orlovsky and Yehuda Salzberg discovered three crucial players that regulate the transcription of the Runx3 gene, and are thus involved in the development of proprioception.
In transcription – the first step by which information encoded in a gene is used to synthesize proteins – such transcription factors as Runx3 bind to a specific DNA sequence called a promoter site that is located near the gene of interest. But many genes are controlled in a complex manner involving additional regulatory elements (REs). These elements are not always near the gene, and can be hard to find.
Few are aware that we have what is sometimes referred to as a ‘sixth sense’ – the position sense
The scientists first employed bacterial artificial chromosomes (BACs) – special gene constructs that harbor large segments of DNA – to express the Runx3 gene and its surrounding regions in transgenic mice. A fluorescent reporter gene was also inserted into the RUNX3 coding region. Groner explains: “Resolving the pattern of expression of these fluorescent reporter-BACs in the transgenic mice revealed a faraway region that was responsible for the specificity of Runx3 expression in TrkC neurons. This region contained the regulatory elements we were looking for.”
“Since Runx3 expression in TrkC neurons has been conserved through the course of evolution, we reasoned that its REs should also be conserved,” says Levanon. “So we compared the RE-containing regions derived from the BAC analysis to analogous regions in the genomes of humans, chickens, frogs and fish, and thereby identified three specific REs that are shared between all these genomes.”
For further proof that these REs, which were named R1, R2 and R3, are indeed regulators of Runx3 expression, the scientists employed the cutting-edge CRISPR/Cas9 technique to delete in mice each RE region separately, or in pairs. The findings, recently published in Genes & Development, showed that deleting only two of the three REs (i.e., R1 and R3) had the same effect as deleting the Runx3 gene itself: Namely, mice depleted of these two small regulatory DNA regions were ataxic.
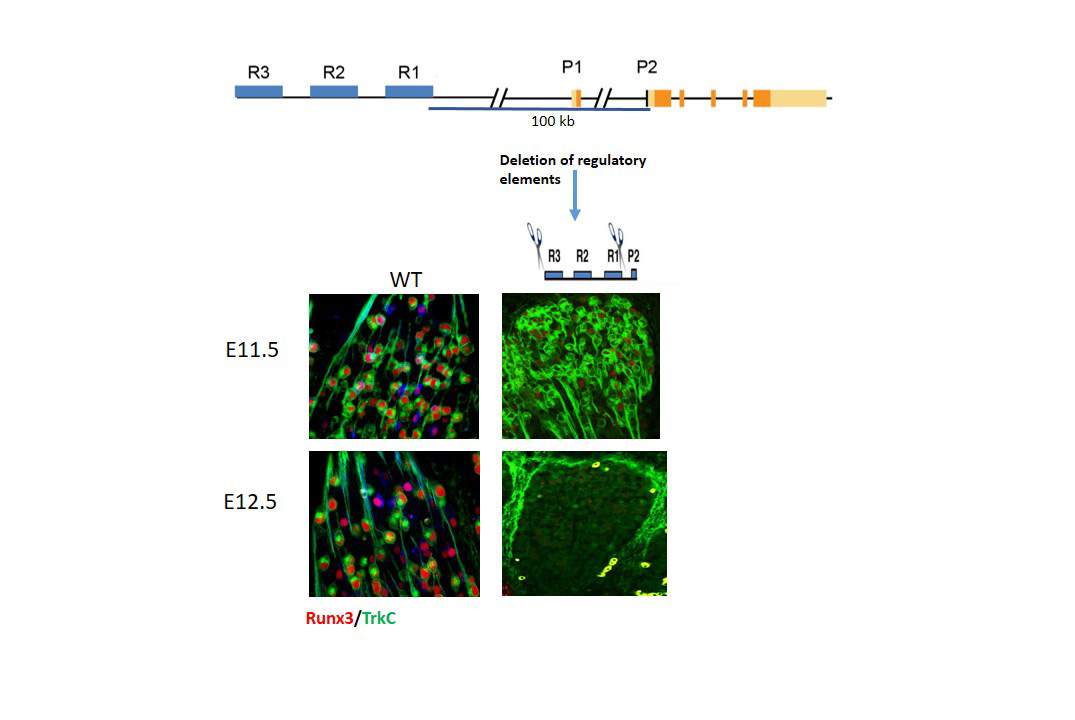
Levanon and Groner and their team found that REs are also active in similar, non-TrkC neurons, in which, rather than promoting Runx3 expression as they do in TrkC neurons, they actively suppress it. They also showed that each RE has a specific role, which in turn, led them to the discovery of TrkC subpopulations. Around embryonic day 11 in mice, R1, R2 and R3 work together to mediate Runx3 expression; but by day 12, an intricate interplay among the three REs mediated Runx3 expression in distinct subtypes of TrkC neurons.
“The results are highly important as there is only a handful of studies that have documented crosstalk between promoters and additional regulatory elements in vivo,” says Levanon. “In the future, it may be possible to use the three regulatory elements that we discovered as a tool to target TrkC neurons in ataxia and other neuropathies involving Runx3.