Are you a journalist? Please sign up here for our press releases
Subscribe to our monthly newsletter:
If we are to stay abreast in our ongoing race against bacteria, with their continuously evolving strategies for thwarting our antibiotics, we desperately need new ways of addressing antibiotic resistance. Research at the Weizmann Institute of Science, which recently appeared in Science, suggests that understanding how antibiotic-resistance genes are “switched on” may point to better ways of overcoming this resistance. After devising a new method to uncover previously unknown RNA switches for bacterial genes, the Weizmann group found new RNA-control switches for genes encoding antibiotic resistance and discovered that these switches are actually “turned on” by the antibiotics themselves.

RNA-control switches, collectively known as “riboswitches,” or “riboregulators,” are short strands of RNA that are attached to messenger RNA. Riboswitches are sensors that turn on or off when they sense a certain level of a particular substance – for example, an amino acid. Riboswitches thus control the genes that are in charge of the production of this amino acid according to the bacterium’s “default setting.”
Some two dozen unique riboswitches had previously been identified, but scientists estimate that hundreds more remain to be discovered. Prof. Rotem Sorek and PhD student Daniel Dar, both of the Weizmann Institute’s Molecular Genetics Department, began their study by developing a method, which they named Term-Seq, for detecting almost all the riboswitches in a cell at once. This method searches for certain sequences at the ends of the RNA strands that tell the scientists a segment is a riboregulator. Bacillus subtilis, a well-studied bacterium, was used to test their system: the researchers identified nearly every known riboswitch in this bacterium and discovered many others that were completely new.
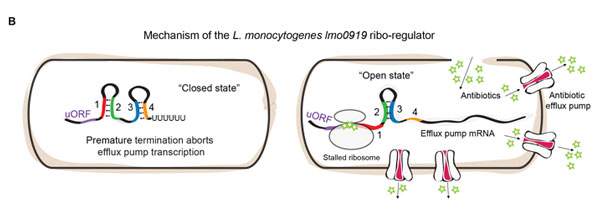
The team then applied this approach to two disease-causing human pathogens: Listeria and Enterococcus. These pathogens, like many others, have genes that enable them to disgorge the antibiotics from the cell, enabling them to survive despite treatment. But how do bacteria know when to turn on their resistance genes? By growing these bacteria in the lab, with or without antibiotics, and applying their method, the researchers found that antibiotic resistance genes were activated by specific RNA-control switches that sense and respond to the antibiotic.
Additional experiments revealed that these particular RNA switches have an “on” and an “off” structure. The “off” structure prevents the production of the protein “pumps” that remove the antibiotics from the cell; the “on” structure, which appears when antibiotics are present, allows the process to proceed. Further research showed that it is the antibiotics’ main target – the ribosome – that decides when to “flip the switch.” When antibiotics bind to the ribosome, they get “stuck” on the RNA switch, forcing the switch to transform itself into the “on” structure, which then activates the resistance gene. In other words, bacteria detect the presence of the antibiotic by monitoring its target; this enables them to recover quickly and remove the threat from their system.
The Weizmann group found new RNA-control switches for genes encoding antibiotic resistance and discovered that these switches are actually “turned on” by the antibiotics themselves
Do other bacteria use similar switching mechanisms to control antibiotic resistance? The team decided to explore the issue with a combination of many different bacteria, and they turned to a nearby source – their teeth. (“You’d be surprised how many bacteria you get by just skipping brushing overnight,” says Sorek.) After collecting the bacteria from the teeth of three team members using a simple toothpick, they again used their approach to search for switches that “turn on” when bacteria are exposed to antibiotics. Indeed, they found a variety of riboregulators in many additional bacterial species, demonstrating that this gene regulation strategy is very common, even in the friendly bacteria we carry around with us daily.
“While antibiotic-resistance genes take many forms, the RNA switches that control them essentially behave in the same way, and these are only found in bacterial cells,” says Sorek. “So they could, in the future, present targets for potential drugs.” The Term-Seq method may be used to understand other types of gene regulation in many different bacteria as well, including the friendly ones in our personal microbiomes. “Once the mechanisms are understood,” says Sorek, “they may also be used in synthetic biology to regulate the production of various proteins. The method is somewhat elaborate, but we think it represents a leap forward for the field, as we can now identify, in one experiment and in a fraction of the time, a good many of the hundreds of riboswitches we know to exist in bacteria.”
Also participating in the study were Maya Shamir of Sorek’s lab, Dr. Noam Stern-Ginossar of the Weizmann Institute’s Molecular Genetics Department and Prof. Pascale Cossart of the Pasteur Institute, France.
Prof. Rotem Sorek's research is supported by the Benoziyo Endowment Fund for the Advancement of Science; the Abisch Frenkel Foundation for the Promotion of Life Sciences; the Leona M. and Harry B. Helmsley Charitable Trust; the Dana and Yossie Hollander Center for Structural Proteomics; the Dr. Dvora and Haim Teitelbaum Endowment Fund; and Mr. Martin Kushner Schnur in honor of Fanny Kushner.