Nothing could be easier than stringing beads – kindergarten children do it every day. Like strings of beads, proteins are made up of small molecular units called amino acids that are linked, one after another, in a long chain. Proteins are assembled by the cellular factories called ribosomes, in which – unlike randomly constructed preschool creations – the amino acids are carefully lined up according to a pre-set pattern laid out in the genetic code. As opposed to the elementary technique for threading a bead on a string, scientists have still not completely answered the question of how the ribosome links the amino acids to one another in the order set down in the genes.
Prof. Ada Yonath of the Weizmann Institute’s Structural Biology Department, working in collaboration with Prof. Lou Massa of the City University of New York and Nobel laureate Prof. Jerome Karle of the U.S. Naval Research Laboratory, recently took some significant steps toward getting to the bottom of this mystery. Yonath has been studying the workings of ribosomes for over 25 years, ever since she, together with scientists from the Max Planck Institutes in Germany, first crystallized them. Ribosomes are composed of a large number of protein molecules loosely bound to giant chains of nucleic acids known as ribosomal RNA. Using a technique called X-ray crystallography, the scientists bombarded the ribosome crystals with X-rays. Some sophisticated mathematical analysis of the scattering of the X-rays bouncing off the crystals revealed the ribosome’s three-dimensional structure in detail.
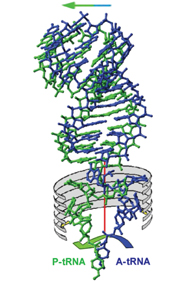
The team’s most recent research focuses on molecular “trucks” that transport the amino acids to the protein production line. Nobel laureate Prof. Aaron Klug and Prof. Alexander Rich (both members of the Weizmann Institute Board of Governors) solved the structure of these “trucks,” called tRNA, two decades ago. Though mainly composed of double-stranded RNA, their ends are single stranded and thus more flexible. All tRNA molecules look misleadingly alike, but they are highly specialized: Each can identify the messenger RNA (mRNA) segment carrying the instructions for a particular amino acid and bring it over to the ribosome. In action, one end of the tRNA molecule attaches to the mRNA as it carries the amino acid bound to its other end to the center of action. There, the protein segment’s bond to the growing chain is manufactured.
Klug and Rich had discovered the structure of these molecules at rest, but it was clear that they undergo significant changes while working. These changes take place so quickly, however, that scientists, until now, had not been able to catch them in the act. Using a molecule that simulates tRNA, Yonath and her team were able to slow down the process and “freeze” the action at various stages
During protein manufacture, the tRNA molecules bind to the ribosome in pairs. One of the pair carries the new amino acid to be incorporated into the growing protein chain, which, in turn, is attached to the second. Once the bond is created, the free tRNA molecule disengages to make way for the next tRNA molecule carrying an amino acid to be attached.
The research team found that while the tRNA molecule’s more stable part moves along with the protein-encoding mRNA, the flexible part – composed of the single-stranded RNA – rotates around a pivot formed by the junction between the two parts. With each rotation, a bond is formed between the amino acid at the end of the tRNA and the growing protein chain.
This rotation is facilitated by the shape of the ribosome. While the ribosome as a whole is asymmetrical, the section where the flexible ends of the “truck” dock is symmetrical, allowing new peptide bonds to form smoothly. By knowing the binding site for the first “truck,” the team was able to calculate the position of the second “truck,” which carries the growing peptide.
Although the team now had a much clearer picture of how the protein machinery works, some finer points of protein chain construction remained elusive. For instance, is the peptide bond formed during the rotation, and if so, at what point does this take place? To answer this question, Yonath, Massa and Karle applied a technique called quantum crystallography, in which the quantum relationships between the atoms in the molecular units are analyzed. The team’s analysis focused on about 50 atoms belonging to the amino acid and the growing protein while both are attached to the tRNA molecule, and calculated how these atoms would be arranged in space if they were completely free to “choose” their positions. Such a “free” arrangement should require the least energy. This calculation showed the scientists that the intermediate state of the reaction is indeed formed during the rotation, and further computations allowed them to trace the energy flow created in the process.
Yonath: “Certain antibiotics work by attaching to the tRNA binding sites on bacterial ribosomes, preventing them from producing necessary proteins. By revealing the details of protein construction in the ribosome, we can advance the development of new antibiotics that will be more effective and possibly attack bacteria that are resistant to existing drugs.”
Prof. Ada Yonath’s research is supported by the Helen and Milton A. Kimmelman Center for Biomolecular Structure and Assembly. Prof. Yonath is the Martin S. and Helen Kimmel Professor of Structural Biology.