Are you a journalist? Please sign up here for our press releases
Subscribe to our monthly newsletter:
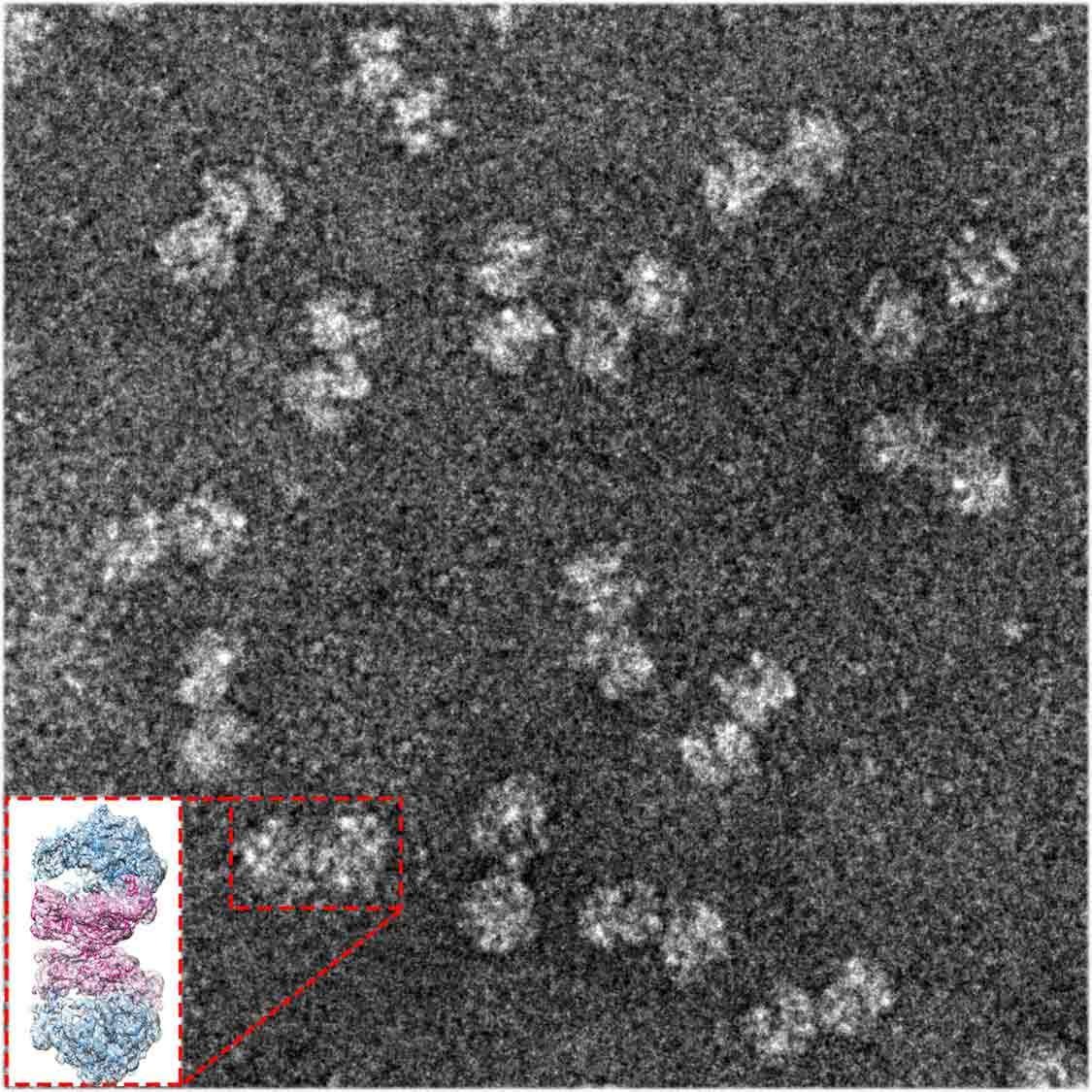
Ribosomes – the protein production machinery of the cell – are a major battleground in the war to defeat harmful bacteria. Many useful antibiotics target the bacteria’s ribosomes mainly by binding to primary functional sites; antibiotic-resistant bacteria invent various means of keeping these drugs from impairing their function by modifying the binding sites. A new study, reported in Nature Communications, might point to a less common way of making some bacterial ribosomes more vulnerable to antibiotics – by keeping them from shutting down naturally.
In bacterial cells, especially those under stress, ribosomes often pair up, attaching to one another through sites on the smaller of their two subunits. When two ribosomes attach in this way, their activity freezes and they are said to go into so-called “hibernation.” This is a temporary situation that can occur, for example, if stress or scarcity force the cell to divert resources from protein production. In some bacteria, under specific conditions, partial hibernation can even be a normal state – possibly slowing down certain ribosome activity. The attachment can reverse itself almost instantaneously, within seconds, and the ribosomes are then “awake” and free to resume their activities. Thus hibernation not only freezes protein production, it preserves the ribosomes in a suspended state.
How do the ribosomes pair up? The process had been documented for the common lab bacterium E. coli, but research had suggested that other bacteria might use alternate mechanisms. These bacteria, which belong to the medical strains called “gram positive,” include the pathogenic Staphylococcus aureus (SA), which can cause everything from mild skin infections to life-threatening illnesses. Understanding the hibernation process in such bacteria, which include multi-drug resistant SA strains for which new antibiotics are urgently needed, may provide crucial insights for the development of future drugs.
To understand how SA ribosomes form pairs, researchers from St. Louis University, Missouri, approached the lab group of Prof. Ada Yonath in the Structural Biology Department of the Weizmann Institute of Science for their unique expertise in unlocking the secrets of the ribosome’s function. This study was led by research student Donna Matzov in Yonath’s lab, who collaborated, in turn, with the electron microscopy group of Dr. Alexey Amunts of Stockholm University, Sweden. Using sensitive purification techniques, the researchers separated all of the ribosomes from cells, and further separated the hibernating ribosomes from the non-paired ones. Matzov brought these to Sweden, where they imaged the samples in exquisite high-resolution three-dimensional detail, and employed state-of-the-art computer algorithms that revealed exactly how a crucial non-ribosomal cellular protein, called hibernating promoting factor (HPF), creates the connection between the two ribosomes. In gram-negative bacteria like E. coli the version of this protein resides near the core of the ribosome. Together with another protein called RMF, HPF sets off a chain of events leading to both the coupling on the outside and the halting of protein production within its active site. Gram-positive bacteria like SA lack RMF, but their HPF protein is twice as long as the E. coli version. “We initially thought that HPF might be something of a hybrid, with an extension that would be analogous to the missing RMF,” says Matzov.
What they found, however, was a completely different mechanism: The HPF of the gram-positive hibernating ribosomes is a relatively long protein with two active sites, one at either end and connected by a flexible loop. Rather than initiating a cascade, its action is direct: One end reaches into the protein production site, blocking activity there, while the other end extends to the outer edge of the small subunit, where it forms a direct bond with the matching HPF protein in the other ribosome. Once this coupling takes place, other parts of the adjoined ribosomes can link into a communications network between the two, thus helping to maintain the ribosomal pairs’ structure and keeping abreast of changes that could let them resume their tasks.

Dr. Anat Bashan of Yonath’s lab, who participated in this research, points out that “the technique that we used is cryo-electron microscopy (cryo-EM), for which this year’s Nobel Prize in Chemistry was awarded. This technique is what enabled us to observe hibernating ribosomes in such detail. Since, in contrast with the traditional crystallography methods, we did not need to crystallize the ribosomes in order to determine their structure, we were able to investigate multi-conformation, dynamic samples.”
Developing a way to prevent hibernation could make the ribosomes more susceptible to antibiotics
“Understanding the function of the HPF protein might be the first step in developing a way to prevent hibernation. This could make the ribosomes more susceptible to antibiotics,” says Matzov. “And since it would target only gram-positive bacteria, many of the friendly bacteria in our bodies might be spared.”
Also participating in this research were Dr. Ella Zimmerman of Yonath’s lab, Dr. Shintaro Aibara of Stockholm University, and Dr. Arnab Basu and Prof. Mee-Ngan Yap of St. Louis University School of Medicine.
Prof. Ada Yonath's research is supported by the Helenand Milton A. Kimmelman Center for Biomolecular Structure and Assembly, which she heads. Prof. Yonath is the incumbent of the Martin S. and Helen Kimmel Professorial Chair of Structural Biology.