Are you a journalist? Please sign up here for our press releases
Subscribe to our monthly newsletter:

Randomness, noise and unpredictability are generally unwanted in machinery, and we try to eliminate them from our mechanical equipment. So we might be forgiven for thinking that the microscopic “machinery” in our bodies − for example, the enzymes that effect chemical changes in other molecules − would have evolved along similar lines. New research in the Weizmann Institute of Science lab of Prof. Gilad Haran, which takes a close-range, high-speed look at the dynamics of an enzyme molecule, suggests that nothing could be further from the truth. On this scale, randomness and noise appear to be basic design principles. These findings, which might lead to a better understanding of how our bodies function at the molecular level and aid in the design of new, more effective drugs or the development of efficient nanomachinery, were recently published in the Proceedings of the National Academy of Sciences (PNAS).
Haran and his group in the Institute’s Chemical and Biological Physics Department have developed techniques to observe motion that occurs on the time scale of microseconds – that is, around millionths of a second – in individual biological molecules. To capture enzyme activity, the researchers used a precise method of observing the structural changes that are involved in binding to a target protein. Multiple enzymes must bring two parts of their structure (called domains) together before their chemical reaction can take place. Among other things, such conformational changes keep the active sites of the enzymes safe until needed and help them locate the exact spot on the target molecules to which they must bind – the substrate. But it had been debated how exactly such a structural change, or domain closure, is related to the biochemical step of the enzymatic reaction.
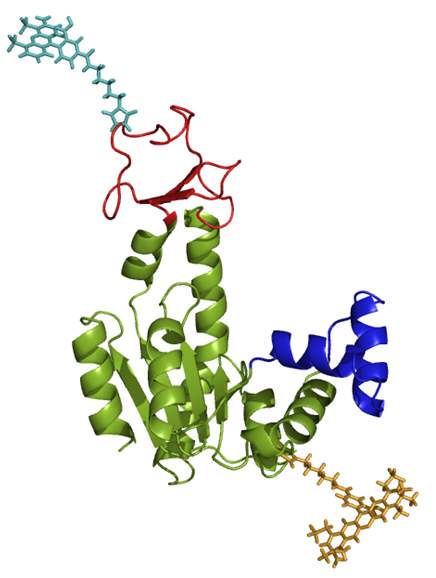
The scientists developed a method in which they isolated single enzyme molecules and chemically attached two tiny, light-sensitive “antennas” – one green, the other red – to each of the ends that come together when the enzyme closes. The green one was tuned so that under a laser beam it would pass its energy to the red antenna if the two were next to each other, but reemit the light if the distance between them was spread out. In this way, the researchers obtained two colors of light – green for the open state of the enzyme and red for the closed state. Detectors so sensitive they are able to record the emission of single photons – to the microsecond – picked up the light signals. “This is really pushing time resolution to the maximum in experiments on single molecules,” says Haran.
The results, he adds, “revealed the molecule frantically clapping open and closed at a fantastic rate.” Cycles of opening and closing took place in less than 100 microseconds. That is about 100 times the rate of the biochemical reaction that ensues. Why would an enzyme close and open again 100 times, rather than closing just once and continuing on to the next step?
Not only have these molecules evolved to work with noise, they have adopted it as a means of operation
“The enzyme may carry out what we think of as a mechanical function,” explains Haran, “but these molecules have none of the properties or conditions we associate with machinery.” First, of course, an enzyme is not rigid, but a floppy molecule moving in a fluid. As opposed to a machine – for example, a piston enclosed in a cylinder and built to repeat an action in the same way every time – a biological molecule is affected by all sorts of random changes in its environment, collectively known as noise. Noise may include other molecules, temperature fluctuations or structural variations in the enzyme itself. Under these conditions, with no GPS, the enzyme molecule must manage to perfectly align its substrates at the active site to get them ready for the chemical reaction. Haran and his team concluded that the enzyme simply continues opening and closing until, at some arbitrary point, it manages to align the substrates correctly so that the reaction occurs. This suggests that randomness plays a more important role in enzyme dynamics than had been conceived.
“In the noisy environments in which enzymes operate, rapid random change may be the most efficient means of carrying out even the most precise, demanding actions. That is, not only have these molecules evolved to work with noise, they have adopted it as a means of operation,” says Haran.
Now that they have applied their ultrafast method to one particular enzyme, Haran and his lab group are using it to study other biological molecules to uncover the fast conformational changes they may undergo. “We think such microsecond dynamics are abundant in biological molecules, and we are hoping to reveal some of the rules and principles that underlie these dynamics,” he says. “Among other things, we believe these principles might be applied to building new types of molecular machines. The Nobel Prize in Chemistry was awarded in 2016 for the ‘design and synthesis of molecular machines,’ which could be used in the development of new materials, sensors and energy storage. But so far, these machines are not capable of doing much more than moving on a track. To create better molecular machines, we might need to think less like conventional engineers, and to embrace noise and randomness.”
Prof. Haran heads the Ilse Katz Institute for Material Sciences and Magnetic Resonance Research and the Nancy and Stephen Grand Research Center for Sensors and Security. Prof. Haran is the incumbent of the Hilda Pomeraniec Memorial Prefessorial Chair.