Are you a journalist? Please sign up here for our press releases
Subscribe to our monthly newsletter:
Our body’s proteins are largely the same as those of apes and monkeys, and the similarities extend even to the less human-like creatures: We share roughly 90 percent of our proteins with mice and 70 percent with fish. But if proteins, and the genes that encode them, have changed relatively little in the course of evolution, other parts of the genome have undergone truly dramatic changes, and these help account for the enormous diversity that has evolved in the animal world. Among the genetic elements that have undergone major changes are lncRNAs (long non-coding RNAs, pronounced link-RNAs), which play regulatory roles, determining which genes are turned on or off, and when.
“Proteins must fold up with great precision in order to perform their functions, and this limits their ability to evolve, whereas lncRNAs are more flexible and looser, so they can apparently undergo major changes without altering the roles they play in the cell,” says research leader Prof. Igor Ulitsky of the Weizmann Institute of Science’s Biological Regulation Department.

Ulitsky and his team have developed a way of detecting similarities between lncRNAs of different species, even those separated by hundreds of millions of years of evolution. Interestingly, many of these lncRNAs have preserved the same function over time. The Weizmann method can be used to identify the parts of a lncRNA molecule that are crucial to its function, including those that may serve as treatment targets in case of disease.
Precisely because they have changed so much over time, lncRNAs of humans and other species at first glance look as if they have nothing in common, certainly not when their continuous genetic sequences are compared. That means that finding parallels between lncRNAs of different species has until now been extremely difficult, in contrast to the ease with which one can correlate proteins of different species with one another.
Ulitsky and his team have developed an algorithm that identifies segments of a lncRNA that have been conserved in evolution, generally a sign that these segments are crucial to that molecule’s function. Among other things, this insight will contribute to the development of effective ways of manipulating the lncRNAs: For example, if a particular lncRNA is known to contribute to disease, identifying its most vital portions will make it easier to block its activity. Moreover, manipulating such vital portions can make it possible to determine which proteins, of the thousands present in the cell, are associated with a particular lncRNA.
If a particular lncRNA is known to contribute to disease, identifying its most vital portions will make it easier to block its activity
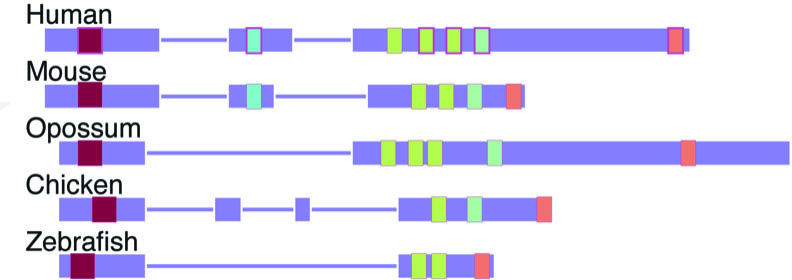
The algorithm, called LncLOOM (for LncRNA Liner Order cOnserved Motifs), regards lncRNA as a weave of beads – important regions of the molecule – strung together on a chain, with less important regions in between. Each “bead” is a short segment, consisting of no more than 15 genetic bases out of the thousands that make up the entire lncRNA. To develop the algorithm, the scientists combined tools from computer science with biological data on lncRNAs in humans and 20 other species, including dogs, opossums, chickens, lizards, fish and sharks.
LncLOOM enabled the scientists to identify lncRNAs that perform the same functions in humans and other species. The closer the species were to one another, the more similar their lncRNAs: They contained the same beads, arranged in the same order. For example, the researchers found that one lncRNA was 10,000 genetic bases long in humans and only 2,000 bases long in fish, but in both species it contained nine matching beads placed in the same order. The team then mined additional data to uncover the biological functions of eight of these beads.
Next, Ulitsky’s team used the algorithm to identify the beads in a lncRNA they called “Chaserr.” In previous work, they had found that Chaserr puts a brake on the activity of Chd2, a gene that is mutated in some people with epilepsy or autism. Because the mutations reduce Chd2’s expression, a potential therapy may involve blocking Chaserr, so it will release its grip on Chd2. It was unclear, however, which parts of the long Chaserr molecule could be effectively blocked. In the new study, the scientists found Chaserr equivalents in 16 different animal species and – by comparing them with the help of LncLOOM – identified this lncRNA’s most essential segments. They then designed a pair of molecules, each of which blocks Chaserr’s activity by binding to one of these segments. When the scientists added the molecules to neuronal cells in a laboratory dish, they observed a potent increase in Chd2. They are currently testing these molecules to see if they alleviate neurological symptoms in a mouse with a Chd2 deficiency that results in a neurological disorder.
The research team included Dr. Caroline Jane Ross, Dr. Aviv Rom, Amit Spinrad, Dikla Gelbard-Solodkin and Dr. Neta Degani, all of Weizmann’s Biological Regulation Department.
Prof. Igor Ulitsky's research is supported by the Benoziyo Endowment Fund for the Advancement of Science; the Sagol Institute for Longevity Research; the Abisch Frenkel Foundation for the Promotion of Life Sciences; the Maurice and Vivienne Wohl Biology Endowment; the Minna-James Heineman Stiftung; the Blavatnik Award; Miel de Botton; Katy and Gary Leff; and the European Research Council.