Are you a journalist? Please sign up here for our press releases
Subscribe to our monthly newsletter:
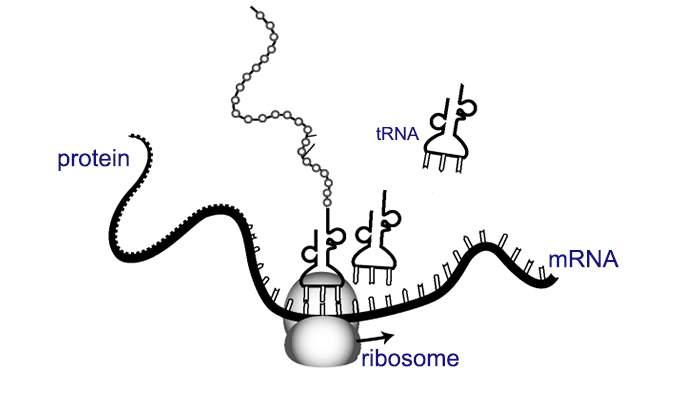
Once the structure of the ribosome – the protein-making machinery of the cell – was solved, new questions about how these machines work arose. For example: How does a strand of messenger RNA (mRNA) recruit the ribosome to start building the protein it encodes? How does the ribosome know where to go? How do viruses hijack this process so that the cell’s ribosomes help them produce new viruses? Research by Weizmann Institute scientists, which recently appeared in Science, sheds some new light on these questions.

Prof. Eran Segal and research student Shira Weingarten-Gabbay of the Weizmann Institute’s Computer Science and Applied Mathematics Department knew that the meeting between ribosome and mRNA molecule is one of the more crucial steps in the process of translating genetic information into proteins. Once the meeting takes place, the ribosome moves down the mRNA strand, reading the genetic information and assembling a protein. The questions for Segal and his group were: What genetic sequences recruit the ribosome to work on the mRNA molecule, and where are these sequences located on the strand?
To find out how this recruitment transpires, the researchers began by creating a library of 55,000 genetic sequences, using a genetic engineering technique they developed for precisely manipulating each sequence to order. Some of the sequences were exact copies of human genetic sequences, some were copied from viruses and others were artificial sequences. They then tested the sequences to see if and how they recruited ribosomes.
To their surprise, a good number of them are actually located near the end of the protein-coding sequence
Up till now, only a few dozen recruitment regions had been identified on various mRNAs; these, unsurprisingly, are located at the beginning of the protein-encoding sequences. But the examination of all 55,000 sequences revealed a new side to this story: The team found thousands of new recruitment regions, and to their surprise, a good number of them are actually located near the end of the protein-coding sequence. This finding, says Segal, changes the way we think about the process of gene-to-protein translation. The mRNA strand, as we know, often bends back on itself to form a loop. If the ribosome is recruited to the “end” on a loop of mRNA, it might, in some cases, begin translating from the end back to the beginning.
Examining these fundamental issues has some interesting biomedical implications, say the scientists. For example, when viruses attack and invade our cells, the recruitment regions on their mRNAs compete with those on our own mRNA, hijacking our cells’ ribosomes for the purpose of producing new viral proteins. If drug molecules could be designed to block these viral recruitment regions, they would be unable to reproduce. The scientists say that the thousands of viral recruitment regions they discovered could be used as a sort of “target library” for designing antiviral drugs.
Also participating in this study were research student Shani Elias-Kirma and Drs. Ronit Nir and Adina Weinberger of Prof. Segal’s group, Dr. Noam Stern-Ginossar of the Molecular Genetics Department, research student Alexey A. Gritsenko of Delft University, The Netherlands, and Dr. Zohar Yakhini of the Technion and Agilent Labs.
Prof. Eran Segal's research is supported by the Crown Human Genome Center, which he heads; the Else Kroener Fresenius Foundation; Donald L. Schwarz, Sherman Oaks, CA; Jack N. Halpern, New York, NY; and Leesa Steinberg, Canada.