When testing a motor only a few molecules in size, determining the “specs” for it can be extremely tricky. Prof. Joel Stavans of the Physics of Complex Systems Department has applied a handy method for measuring the specifications of one such naturally occurring motor, a protein complex called RuvAB that forms part of the cell’s DNA back-up repair service.
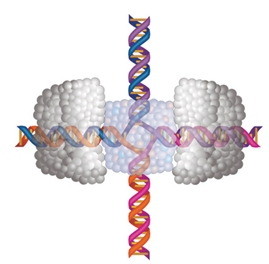
To bypass defects, two identical double strands of DNA may become intertwined at a point called a Holliday junction (see diagram). The RuvAB complex then clamps onto either side of this junction, extruding DNA out to the sides. As two arms grow at the expense of the other two, the junction shifts down the strands, bypassing the glitch so the DNA can be copied correctly.
Stavans, together with Ph.D. student Roee Amit and Dr. Ofer Gileadi of the Molecular Genetics Department, devised a Holliday junction with two very long arms and two short arms. They attached a bead to one of the long arms, while the opposite arm was anchored to a surface. Though only a few millionths of a meter in breadth, the bead was about a thousand times wider than the DNA, allowing the scientists to observe it under an optical microscope. They then added RuvAB complexes. As the RuvAB motor worked, it progressively shortened the long arms, dragging the bead closer to the anchor point. By the bead’s movement, the team worked out the speed of the motor as well as the duration of its activity.
To their surprise, the experiment also revealed that this nanomotor even changes gears as it works, varying the speed at which it moves the junction point along the strands.
Prof. Stavans’ research is supported by the Clore Center for Biological Physics; the Fritz Thyssen Stiftung and the Rosa and Emilio Segre Research Award.