Are you a journalist? Please sign up here for our press releases
Subscribe to our monthly newsletter:

Prof. Yardena Samuels of the Weizmann Institute’s Molecular Cell Biology Department is a part of the Cancer Atlas group that recently produced the melanoma genome map. Hundreds of researchers from Australia, the US, Canada, Russia, Germany, Italy, Poland, China and Korea participated in the effort. “This has been the most in-depth mapping yet,” says Samuels. After a careful selection process, 333 melanoma samples were included in the study, and the screening was conducted using six different platform technologies, going beyond the bounds of simple gene sequencing to explore how the various genes are expressed, how they interact and which proteins they produce.
Each sample required a second, non-cancerous sample from the same patient for comparison, and several of the samples afforded comparisons of genes from the patients’ early tumors to those that had metastasized. In addition to the sequencing of protein-coding genes, some of the samples had their entire genome sequenced, which could prove in the future to be an “unexplored goldmine of information on what makes cancer tick,” says Samuels. RNA and microRNA, as well as protein expression assays, were included. The latter screens added further dimensions to the gene map – creating a “landscape” that can help researchers understand not just the genes, but the pathways, intersections and cancer-causing diversions associated with them.
“The study was intensive, and it has paid off,” says Samuels. For one thing, it showed, for the first time, that melanomas can be divided into four distinct groups, according to a main mutation. Now melanoma researchers will be able to focus on understanding exactly how the different mutations lead to cancer, and physicians may eventualy gain better tools for diagnosing the disease and tailoring treatments to individual cases. The first type, occurring in a mutation “hotspot,” is known as BRAF, and it tends to appear in younger people, in whom the cancer is fairly aggressive. The second is known as RAS. BRAF and RAS protein products lie in the same pathway so that BRAF and RAS mutations are mutually exclusive: A patient will have one or the other, but not both. The third is called NF1, and this mutation was found in older patients. The fourth group, called “triple wild type,” had none of the three most common mutations.
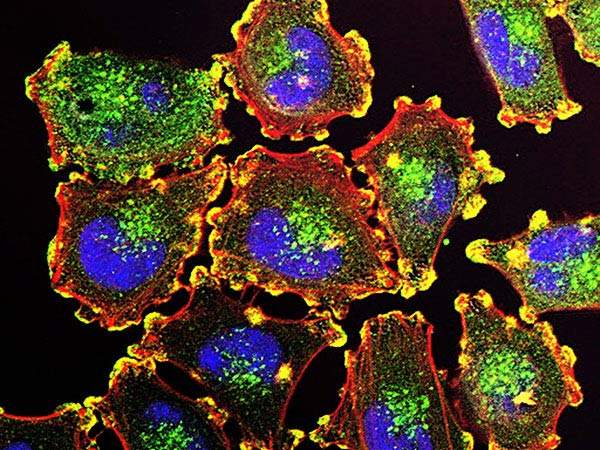
In addition, the in-depth analysis held a surprise finding. RNA and protein expression tests revealed the infiltration of immune cells called lymphocytes into the tumors. These turned out to be highly correlated with the patient’s prognosis – the more lymphocytes, the better their chances of surviving, regardless of the genetic signature of their melanoma cells. This finding has interesting implications for the field of cancer immunotherapy, in which a patient’s immune cells are “trained” to fight the cancer. It suggests that even upping the numbers of particular immune cells could have a positive effect, and it could help identify those patients who might benefit from various new immunotherapies.
Another area of research arising from the new cancer genome that could bear fruit in the near future is a sort of “matching” of existing drug compounds to the genetic profiles. For example, a comparison of genomes from different cancers shows that the genetic mutation pattern of one type of melanoma is similar to that of a form of a brain cancer called glioblastoma. That means that drugs already on the market for glioblastoma might have an effect on melanoma as well.
Separating the drivers from the passengers
Samuels says she intends to continue adding detail to the map – her lab has its own bank of melanoma cells, and she is creating an expanded database of melanoma genomes. This melanoma cell bank is an important resource for her lab, where she and her group are working to figure out which of the many mutated genes drive the development of melanoma, which help the drivers "steer," and which are just “passengers.” Experiments on cell lines from the bank, for example, enable the group to investigate the effects of individual genes, and they are going after are suspected drivers, as well as the "helpers."
In parallel, she is beginning to explore the pathways – the series of biological interactions that underlie each action in the cell – in two of the melanoma subgroups. “If the mutation in a gene, for example, a tumor suppressor, causes a loss of function, you can’t fix it by blocking the gene – the problem is that the gene is not working in the first place,” she says. “But if you follow the pathway, you are likely to find other genes that present targets for turning the pathway around, farther down the line.
“We are entering a new era of precision medicine in melanoma, in which physicians will aim to determine the personal profile of each cancer and tailor the treatment accordingly,” she says.
Prof. Yardena Samuels' research is supported by the Ekard Institute for Diagnosis, which she heads; the Henry Chanoch Krenter Institute for Biomedical Imaging and Genomics; the Laboratory in the name of M.E.H. Fund established by Margot and Ernst Hamburger; the Louis and Fannie Tolz Collaborative Research Project; the Dukler Fund for Cancer Research; the European Research Council; the De Benedetti Foundation-Cherasco 1547; the Peter and Patricia Gruber Awards; the Comisaroff Family Trust; the Rising Tide Foundation; the estate of Alice Schwarz-Gardos; the estate of John Hunter; the Estate of Adrian Finer. Prof. Samuels is the incumbent of the Knell Family Professorial Chair.