A protein starts out as a long “beaded necklace” of amino acids; the sequence of the beads determines the protein’s primary structure. But it is not until it matures into a folded 3-D structure that it becomes functional – ready to carry out one of the many vital tasks of proteins in the body, from providing mechanical support to moving such materials as oxygen to catalyzing other proteins. Scientists have spent decades developing tools and techniques to decipher the structure of proteins.
Sometimes, however, the journey is just as important as the destination: How do proteins get from their unfolded to their folded state? In addition to advancing basic biological research, understanding the mechanisms of protein folding could have implications for disease research, given that a number of diseases, among them Alzheimer’s, involve misfolded proteins.
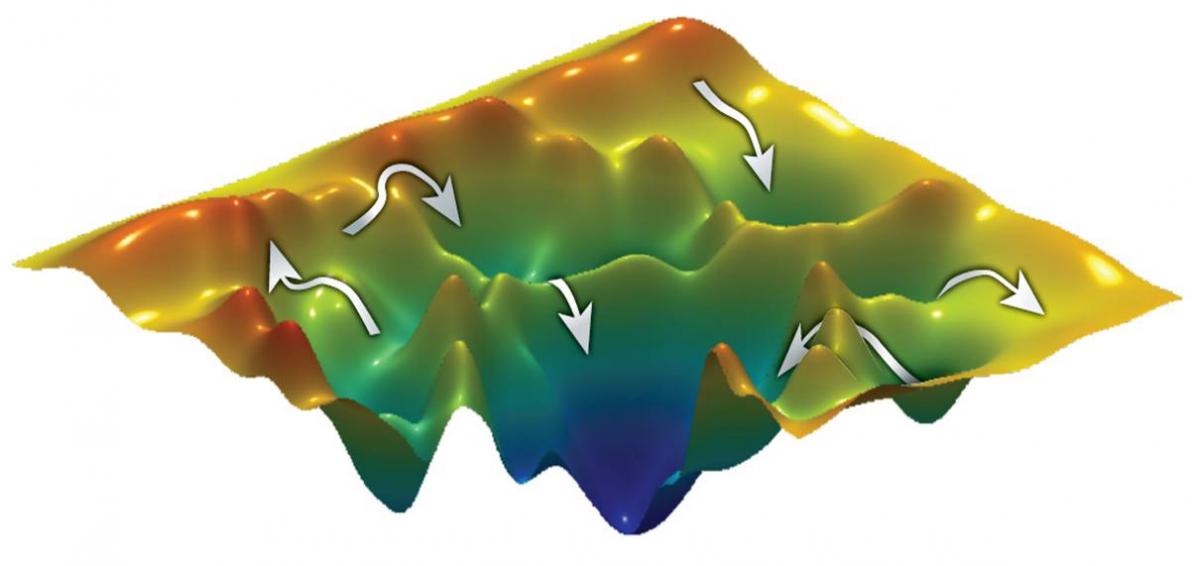
Analyzing the folding mechanisms of small proteins is relatively simple: Just one step is required to get from the unfolded to the folded state. But larger, more complex proteins are a different story. “Analyzing the folding mechanisms of larger proteins is a bit like cartography,” explains
Prof. Gilad Haran of the Weizmann Institute’s Chemical Physics Department (Chemistry Faculty). “Many routes may lead to the same point, some tracks being straightforward and ‘easy-going,’ while others have to cross strenuous terrain to reach the same destination. By mapping proteins, we hope to discover whether these have a fixed number of ‘pathways’ – intermediate conformational changes – leading to their final folded structure. And if this is the case, how many? Do they have to pass through all possible conformations or can they take a ‘shortcut’ and bypass some steps? Do they have a preferred ‘route,’ needing to follow a specific sequence? Or do external conditions, such as temperature, affect their behavior?”
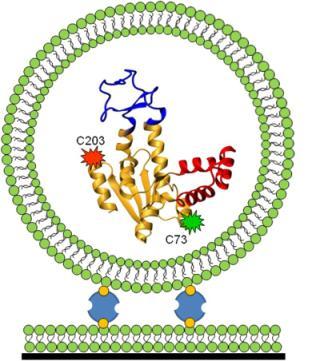
To address these questions, Haran developed a novel technique. Together with his team, including Ph.D. students Menahem Pirchi, Sharona Sedghani Cohen and Nir Zohar; former Ph.D. student Guy Ziv; postdoctoral fellow Dr. Inbal Riven and staff scientist from the Chemical Research Support Department Dr. Yoav Barak, Haran took a protein (adenylate kinase) and attached fluorescent dye molecules at two different points along the length of the protein’s chain. When the two dyed points are far apart (as in the case of an unfolded protein), green light is emitted, and when they are in close proximity (when the protein is folded), red light is emitted. By measuring the dyes’ fluorescent properties, the scientists were hoping to map the “coordinates” of the tagged beads and determine how many intermediate states it took for the protein to get from its unfolded to its folded state.
Here, the team of protein mapmakers ran into a few snags. For one, it is difficult to track down freely roaming protein molecules long enough to analyze them. Haran devised a trap: He encapsulated each protein molecule within a vesicle and tethered it to a surface, thus immobilizing the protein so that measurements could be carried out. Another catch was that the fluorescent dye loses its fluorescence within a few seconds, not providing enough time for the protein folding event to be traced in its entirety. To tackle this problem, the team took thousands of single-protein molecules and analyzed short traces of folding events, each one starting at a different point in time. At this stage, the researchers resembled a biographer whose pages have suddenly been blown by a gust of wind; to return things to the proper order, they developed a statistical analysis that "stitched" together the short sequences of folding events in their correct chronological sequence.
The scientists discovered, as recently reported in
Nature Communications, that for the protein they analyzed, there are a total of six possible states that lead to its 3-D structure. The route this protein chose depended on such external factors as temperature or chemicals: The higher the chemical concentration, for example, the more likely the protein was to opt for the longer, more arduous route – folding into all six of the possible conformations in sequential order before reaching its destination. At lower concentrations, the protein was able to follow easier routes, taking shortcuts that bypassed different conformational states while still ending up at the same folded structure.
Despite the challenges in designing a method for following the activities of individual protein molecules (as opposed to standard methods that average the behavior of thousands of molecules), the techniques the team developed enabled them to obtain a highly detailed picture, with all its variations, of protein folding. They now plan to explore different protein "landscapes" to gain a deeper understanding: Is there a general folding rule that applies to all proteins? And what are the forces that guide a molecule to fold?
Prof. Gilad Haran’s research is supported by the Carolito Stiftung. Prof. Haran is the incumbent of the Hilda Pomeraniec Memorial Professorial Chair.