Have humans beaten nature at its own game? The attempt required a team of scientists using some of the most advanced computer-aided design and biological engineering techniques available, as well as one very ancient technique – evolution. Recently scientists created a new enzyme that performs a function unknown in nature. Compared to the millions of years nature takes to come up with a new enzyme, the scientists managed this feat in hardly any time at all.
Enzymes are biological catalysts – dynamic molecular machines that initiate chemical reactions in the body and speed them up millions of times. Each of the tens of thousands of enzymes in the human body is constructed of a long string of amino acids (the building blocks of proteins), and their sequence determines the intricate three-dimensional shape they assume as the string folds in and around itself. Whereas nature fashions enzymes to fit different functions by adapting basic designs over millions of years, the scientists started from scratch with a problem: How to speed up a common chemical reaction in which a proton (a positively-charged hydrogen atom) is removed from carbon, breaking the tough carbon-hydrogen bond. No existing natural enzyme performs this task with the particular substrate the team applied.
The team began with the design of the active site – a string of amino acids responsible for the chemical reactions that take place. They then turned their attention to the rest of the enzyme – its backbone. This entailed generating a sequence for the 200 amino acids in its protein structure. While in theory there are a nearly infinite number of ways to arrange the 20 types of amino acid into strings of 200, in practice only a limited number can yield a structure, and thus a mode of action, capable of performing the task.
To identify possible sequences for their enzyme, Prof. David Baker of the University of Washington, Seattle, used novel computational methods to scan tens of thousands of computer-designed sequences. Eventually he and his team identified 60 that could potentially facilitate the chemical reaction, and these he tested further to see if they would work in biological systems.
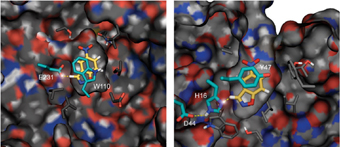
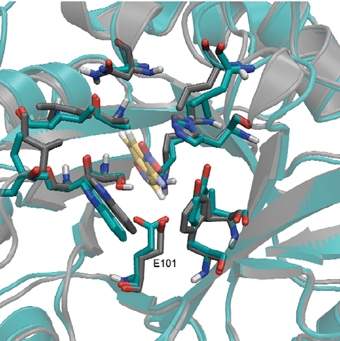
Of the 60 sequences tested, eight showed signs of biological function and only three made it to a “final round” of the scientists’ challenge by showing a detectable level of activity. At this stage, the enzymes had moved from the computer “drawing board” to the bioengineering lab. Drs. Orly Dym and Shira Albeck of the Weizmann Institute’s Structural Biology Department solved the structure of one of the final contenders, confirming that the structure of the enzymes created was essentially identical to the predicted computational design.
Yet even with the benefit of the most advanced computer and bioengineering technologies, the winning enzymes could not compete with the speed and efficiency of their naturally evolved counterparts. At this point, Prof. Dan Tawfik and research student Olga Khersonsky of the Biological Chemistry Department entered the picture. In a twist on natural evolution, they have found that the principles of random mutation and natural selection can be applied in the lab. Test-tube grown enzymes are mutated, and the most efficient ones are selected to undergo further rounds of mutation and selection.
At the end of only seven rounds of test-tube evolution, the enzymes’ efficiency had increased 200-fold over those created from the computer design template, and the new enzyme-assisted chemical reaction rates were a million times those taking place with no enzyme at all. While the designed/evolved enzymes are still much slower than their natural counterparts, a comparison of the evolved enzymes to their computer-designed ancestors showed how adjustments to the enzyme structure can increase its efficiency: One near the active site improved the reaction rate, while others increased the molecules’ flexibility, allowing the enzyme to detach faster upon completing a step in the reaction.
“The combination of computational design and molecular in vitro evolution opens up new horizons in the creation of synthetic enzymes,” says Tawfik. “Reproducing the breathtaking performance of natural enzymes is a daunting task; but thanks to this research, we have gained a better understanding of the structure of enzymes as well as their mode of action. This, in turn, will allow us to design and create enzymes that nature itself has not ‘thought’ of. The possibilities are nearly endless: neutralizing poisons, developing medicines and future applications that we ourselves have not yet imagined.”
Prof. Dan Tawfik’s research is supported by the J&R Center for Scientific Research; the Wolfson Family Charitable Trust; the Jack Wolgin Prize for Scientific Excellence; Mr. and Mrs. Yossie Hollander, Israel; Rowland Schaefer, New York, NY; and the estate of Fannie Sherr, New York, NY.