Having received the command, the computer starts churning out information, systematically filling in tables and charting graph after graph. This may sound like a dream research scenario, yet as most scientists would quickly point out, it's just the beginning. The true challenge is to make sense of the data.
Recent developments in genetic research are a good example. New genetic research technologies, such as DNA chips, enable scientists to evaluate simultaneously tissue samples from several patients, expressing thousands of genes. However, deciphering the vast amount of resulting information consisting of anything from 100,000 to 1,000,000 genetic "figures," requires highly sophisticated data processing tools.
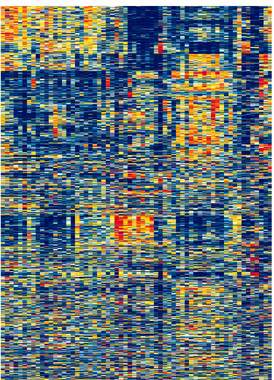
Addressing this and similar challenges may soon be easier thanks to Prof. Eytan Domany of the Weizmann Institute's Physics of Complex Systems Department and doctoral students Gad Getz and Erel Levine. The team has designed a unique mathematical system for analyzing genetic data based on a computer algorithm that "clusters" information into relevant categories. The algorithm searches simultaneously for clusters of "similar" genes and patients by evaluating the gene expression of tissue samples. (A gene's "expression" refers to the production level of the proteins it encodes.)
Reported in the Proceedings of the National Academy of Sciences (PNAS), the algorithm's most powerful feature is that it mimics unassisted learning. Unlike most automated "sorting" processes, in which a computer must be informed of the relevant categories in advance, the algorithm is analogous to human intuition (such as the ability to intuitively categorize images of animals and cars into proper classes). When given a clustering task, it analyzes the data, computes the degree of similarity among components, and determines its own clustering criteria.
The new method makes use of a previous application formulated by Domany and his colleagues, based on a well-known physical phenomenon. When a granular magnet such as a magnetic tape is warm, its grains are highly disorganized. But upon cooling down, the magnet's grains progressively organize themselves into well-ordered clusters. Using the statistical mechanics of granular magnets, Domany created an algorithm that can look for clusters in any data.
When applied in a cancer study using DNA chips, the new algorithm proved highly effective, evaluating roughly 140,000 figures representing the cellular expression of 2,000 genes from 70 subjects. The algorithm categorized tissue samples into separate clusters according to their gene expression profiles. For example, one cluster consisted of cancerous tissues, while another contained samples from healthy subjects. The new method also distinguished among different forms of cancer and demonstrated treatment effects, picking up differences in the gene expression of leukemia patients that had received treatment versus those that had not. Finally, one of the algorithm's most promising features is that it enabled researchers to pinpoint a small group of genes from within the 2,000 examined that can be used to accurately distinguish among cellular cancerous processes.
In a sense, however, applying the new algorithm to DNA chips is only a start. The new algorithm's inherent clustering capacity makes it invaluable for use in data-heavy scientific and industrial applications. It may be used to analyze financial information and MRI data in brain research, or to perform "data mining," the process by which specific details are culled from the world's huge and ever-growing data banks, such as those generated by the international Human Genome Project.
The Institute's technology transfer arm, Yeda Research and Development, has issued a patent application for the algorithm.