When you run, your body is undergoing all sorts of metabolic changes: A slew of substances produced from head to toe increase circulation and breathing rates, cause muscles to take up and burn sugars, open the pores in the skin and induce fatigue. Yet the changes that can presently be measured during exercise – for example, one’s heart rate – are few. If the levels of these substances could be tracked as they are released and go to work, a much more complete picture of an individual’s metabolic processes could be obtained. Since metabolic malfunctions are involved in many diseases – from circulatory and cardiac diseases to diabetes and many cancers – such a method could be a highly useful diagnostic tool to understand health and disease.
Prof. Lucio Frydman and his coworkers in the Chemical Physics Department recently developed a new method based on MRI scanning that can find these substances (metabolites) and track their levels in real time – for instance, as muscles are stimulated.
This method is related to widely used functional MRI technologies such as those used in brain research, but it entails a significant departure from them. Standard MRI, explains Frydman, relies on basic physical properties of the nuclei of the water molecules in the body. At the heart of an MRI exam is the scanner’s strong magnet, which polarizes the water’s protons. These nuclei behave as tiny compass needles aligning with the MRI magnet, enabling the subsequent detection of their collective “nuclear symphony.” Yet at room temperatures, only around one in 100,000 water molecules is actually aligned – or polarized – by the scanner, while the rest basically stay “silent” throughout the “symphony.” This tiny proportion of polarized protons is adequate for reporting on water, since it is present in very high concentrations in tissues. But it cannot capture the low concentrations of metabolites as they are produced in real time – that is, when tissues or organs become involved in a particular task.
The new method that Frydman, together with Drs. Avigdor Leftin and Tangi Roussel in his lab, developed, is based on a technique that they and others have been working on over the past several years, called nuclear hyperpolarization. Using new physics-based concepts and experiments to transform a natural alignment of only one in 100,000 molecules into that of around one in every five molecules, hyperpolarization creates a phenomenal increase in the “symphony’s volume.”
The method involves taking the wanted metabolite, cooling it to near absolute zero – a temperature at which magnetism is strongest – and using free electrons to further align the compass-like nuclei of interest. Combined, these procedures lead to intense metabolic signals, but under conditions that are incompatible with MRI investigations in living organisms. To get around this incompatibility, the hyperpolarization procedure is carried out in a special device positioned right next to the MRI scanner. When the substance to be analyzed has been sufficiently magnetized, the compound is rapidly heated to room temperature and injected into the organism. The nuclei retain their alignment long enough to produce a single, but highly intense, image in the MRI.
Though involved, the method could lead to a highly sensitive tool for detecting the functional MRI response of individual metabolites. To demonstrate, the team focused on muscle stimulation. The skeletal muscles – those you use to run, for example – can be broadly divided into two classes, depending on whether they are rich in fast-twitch or slow-twitch fibers. Fast-twitch muscles – the kind that help you sprint – produce lactate as a byproduct; while slow-twitch muscles – the kind your body uses for endurance running – make much less of this metabolite. Athletes who excel at either short races or marathons often have more of one kind of muscle fiber than the other.
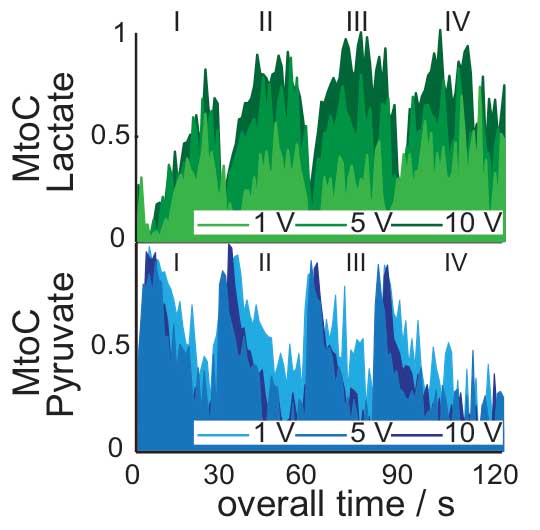
To image the rate of lactate production, Frydman and his coworkers focused on pyruvate: a harmless tracer that the body would naturally use – particularly under certain exercise conditions – to create lactate. Using nuclear hyperpolarization together with custom-made MRI sequences that could collect multiple rapid, simultaneous images of individual metabolites in the muscles, they followed the processes taking place in the exercising thigh of a living mouse. The hyperpolarized tracer was injected via the mouse’s tail and, as the rate of exertion changed (controlled by mild electrical stimulation), the researchers looked at the signature of the pyruvate tracer, which was metabolized into lactate and other byproducts. Sure enough, the team identified peaks associated with lactate production as the muscles were stimulated; the imaging techniques revealed more rapid rates of lactate production in the fast-twitch than in the slow-twitch fibers.
This study, which appeared in
PLoS ONE, is a clear demonstration that the combination of advanced hyperpolarization methods and customized magnetic resonance techniques can extend the technology far beyond today’s functional MRI, to one based on metabolic signatures. This technique might open many doors to diagnosis, enabling clinicians or researchers, for example, to observe what happens in the body after glucose enters the bloodstream, how a tumor takes in nutrients or what happens to our bodies when we are frightened.
Prof. Lucio Frydman’s research is supported by the Ilse Katz Institute for Material Sciences and Magnetic Resonance Research; the Helen and Martin Kimmel Award for Innovative Investigation; the Helen and Martin Kimmel Institute for Magnetic Resonance Research, which he heads; the Adelis Foundation; the Mary Ralph Designated Philanthropic Fund of the Jewish Community Endowment Fund; Gary and Katy Leff, Calabasas, CA; Paul and Tina Gardner, Austin TX; and the European Research Council.
Caption
Type your text here