Are you a journalist? Please sign up here for our press releases
Subscribe to our monthly newsletter:
When Dr. Shiran Barber-Zucker joined the lab of Prof. Sarel Fleishman as a postdoctoral fellow, she chose to pursue an environmental dream: breaking down plastic waste into useful chemicals. Nature has clever ways of decomposing tough materials: Dead trees, for example, are recycled by white-rot fungi, whose enzymes degrade wood into nutrients that return to the soil. So why not coax the same enzymes into degrading man-made waste?
Barber-Zucker’s problem was that these enzymes, called versatile peroxidases, are notoriously unstable. “These natural enzymes are real prima donnas; they are extremely difficult to work with,” says Fleishman, of the Biomolecular Sciences Department at the Weizmann Institute of Science. Over the past few years, his lab has developed computational methods that are being used by thousands of research teams around the world to design enzymes and other proteins with enhanced stability and additional desired properties. For such methods to be applied, however, a protein’s precise molecular structure must be known. This typically means that the protein must be sufficiently stable to form crystals, which can be bombarded with X-rays to reveal their structure in 3D. This structure is then tweaked using the lab’s algorithms to design an improved protein that doesn’t exist in nature. But if the original protein cannot even be produced in the lab or is too fragile to form crystals, as is the case with versatile peroxidases, such attempts at improvement can run into a dead end.

Barber-Zucker nonetheless took a chance on the prima donna enzymes, and her timing was uncanny. Since the 1980s, attempts have been made to get around the need for crystallization by predicting a protein’s 3D structure from its DNA sequence, but for complex proteins such as peroxidases, these predictions were unreliable. Yet in late 2020, several weeks after embarking on her project, Barber-Zucker’s predicted enzyme structures suddenly looked surprisingly reliable. It turned out that at just that time, Google’s company DeepMind and several university research teams had improved artificial intelligence (AI)-based structure prediction methods to the point where they had become highly accurate. This proved to be a game changer: The approach has led to predicted models that are nearly as accurate as those obtained experimentally with crystallography.
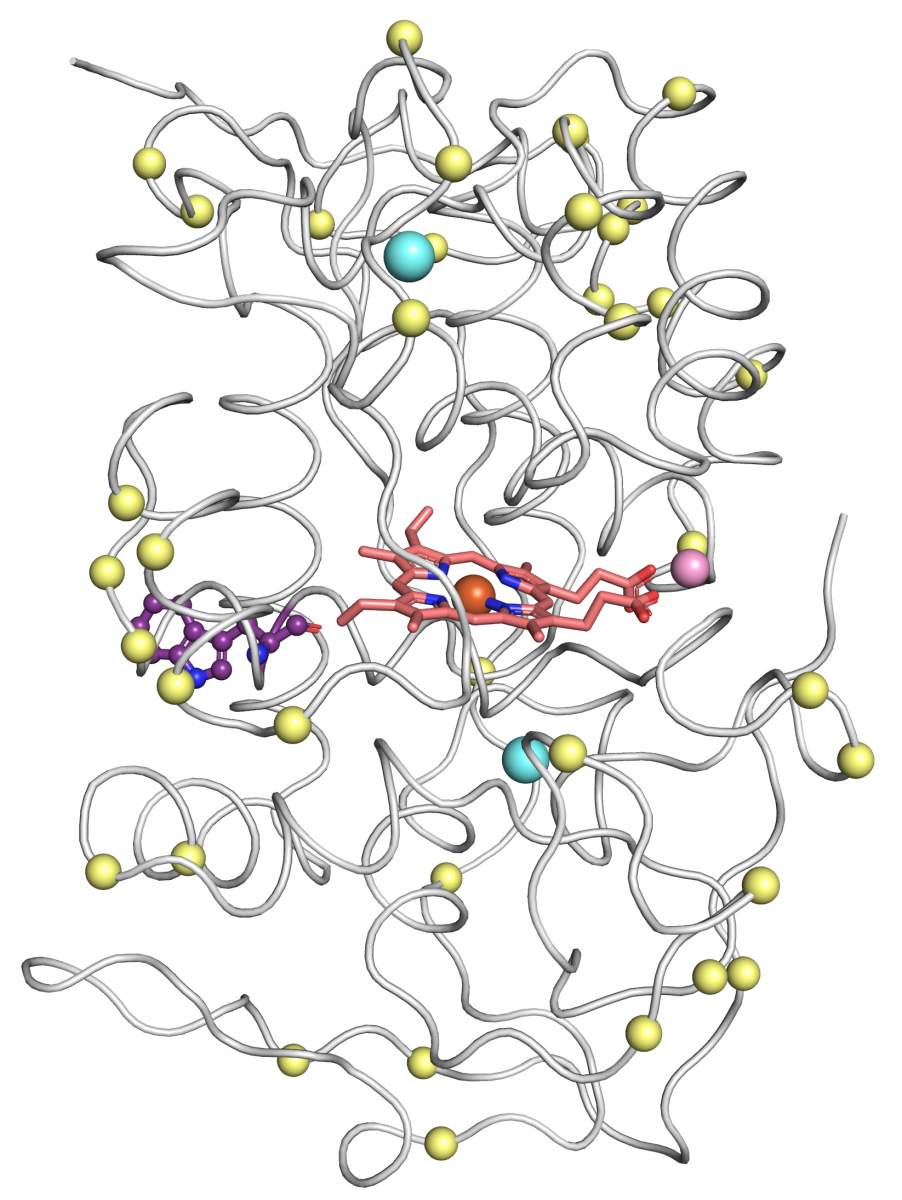
Armed with the new structures, Barber-Zucker, together with colleagues – Vladimir Mindel and Jonathan J. Weinstein, research students in Fleishman’s lab, and Prof. Miguel Alcalde and Dr. Eva García Ruiz of the Institute of Catalysis in Madrid – achieved the previously unthinkable. Only one enzyme in the versatile peroxidase family had already been described structurally by researchers, and that project had taken a team of experts about a decade. Now, within less than six months and without any prior expertise in wood-degrading enzymes, Barber-Zucker and colleagues managed to design, produce and analyze stable variants of three versatile peroxidases whose original versions could not, in the past, have been produced in the lab. The scientists used AI-based 3D models as their starting point. They applied to these models an algorithm created in Fleishman’s lab called the Protein Repair One Stop Shop, or PROSS, which designs an altered protein on the computer to improve its properties on demand.
"In previous protein design studies, our first question was, ‘Do we have a structure of the protein we want to focus on?’ But now this question has become irrelevant; that’s a true turning point.”
This combined approach opens an enormous range of opportunities. “Millions of potentially valuable proteins that once could not have been accessed biochemically are now within reach for research and for use in biomedicine and chemistry,” Fleishman says. He is referring to the fact that 3D structures have been solved experimentally for less than 0.05 percent of the millions of natural proteins whose DNA sequence is known, and that about half of all proteins in nature cannot be effectively expressed and tested in the lab. “These proteins are the dark matter of biology – scientists have no way of accurately determining what they do. In previous protein design studies, our first question was, ‘Do we have a structure of the protein we want to focus on?’ But now this question has become irrelevant; we can manage with a structure or without, and that’s a true turning point.”
Drug design is one area that could immediately benefit from this advance. For example, antibodies created in lab animals must be adapted to humans before they can be used in a clinical setting – a laborious process that involves crystallization and altering numerous regions of the animal molecule. The new advance is expected to make this and other antibody engineering processes much more efficient and effective.
Environmental applications, the original rationale for this study, are another promising avenue. Wood-degrading enzymes could, for example, be adapted for recycling tough agricultural waste. Instead of burning such waste or dissolving it with polluting chemicals, as is often done today, it may be possible to break it down, using versatile peroxidases, into sugars that can be fermented into biofuel. Farmers would then be able to perform recycling in small bioreactors.
The enzymes could also be designed to degrade environmental pollutants. In fact, Barber-Zucker has already shown that her improved enzymes can attack a particularly stubborn polluting dye. She also found that each of the three improved enzymes exhibited a different activity in the lab, and that each specialized in degrading different wood components, which suggests that they may act synergistically. Importantly, all three enzymes proved remarkably stable and resistant to heat, an essential feature for their use in industry. Barber-Zucker now aims to develop an enzyme “cocktail,” in which a dozen different enzymes, including her versatile peroxidases, will work synergistically to break down waste wood into biofuel or other useful materials.
And what about her vision of recycling hard plastics using these enzymes? “That’s still a dream, but one that may become a reality in the near future,” she says.
DNA sequences of some 300 million natural proteins – derived from marine viruses, soil bacteria, plants and animals – have been deposited in public databases. Fewer than 150 thousand proteins have been crystallized and described structurally.
Prof. Fleishman’s research is supported by the Dr. Barry Sherman Institute for Medicinal Chemistry; the Nancy and Stephen Grand Center for Sensors and Security; the Schwartz/Reisman Collaborative Science Program; the Sam Ousher Switzer Charitable Foundation; the Dianne and Irving Kipnes Foundation; Ms. Darlene Switzer-Foster and Mr. Bill Foster; Carolyn Hewitt and Anne Christopoulos, in Memory of Sam Switzer; the Milner Foundation; and the Ben B. and Joyce E. Eisenberg Foundation.