To divide or not to divide: This is one of the most crucial decisions in the life of a cell. A new Weizmann Institute of Science study, conducted in collaboration with Danish researchers, has revealed a previously unknown mechanism that is involved in the control of such decisions in both healthy and cancerous cells.
The findings, reported recently in
Cell, provide an entirely new feature by which dividing and non-dividing cells can be told apart – a discovery that may, in the future, help advance the diagnosis, prognosis and perhaps even therapy of cancer.
Until now, the existence of more than 60 genetic three-letter synonyms, each having a corresponding tRNA, was not thought to have a major bearing on cell division. The ratios of different tRNAs were thought to be constant throughout the life of each cell, and in any event deviations or fluctuations in these ratios were not known to have particular biological significance.
Prof. Yitzhak Pilpel of the Weizmann Institute’s Molecular Genetics Department decided to look into the potential role played by the relative amounts of different tRNAs in the cell when he learned that Danish researchers had compiled an enormous database of genomic information from more than 400 people, including some 300 cancer patients, which contained a large volume of tRNA data. Pilpel’s intern Dr. Hila Gingold used DNA chip technology and mathematical algorithms to analyze the data, working together with Dr. Disa Tehler and other members from the team of Prof. Anders Lund of the University of Copenhagen, as well as other researchers from Denmark, the Netherlands, Switzerland and the United States.
Division signs
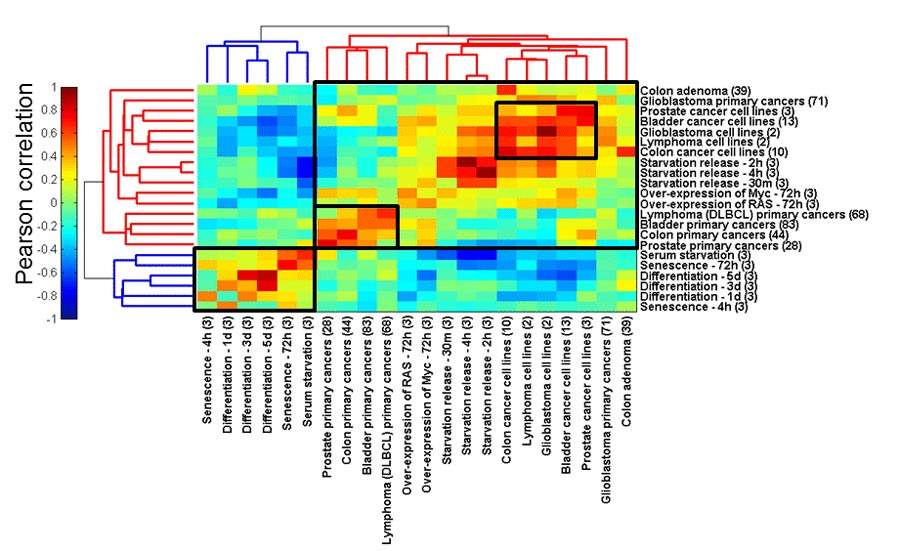
The results were so striking they surprised even the study authors themselves. The analysis revealed that two distinct subsets of tRNAs are present in the cell depending on whether it is dividing or not. For example, dividing cells tend to have large amounts of the tRNA that translates the GAG synonym, whereas non-dividing cells have a preference for the tRNA associated with the GAA synonym. As a result, dividing and non-dividing cells have discernible tRNA signatures – that is, characteristic amounts of all the different tRNAs.
In other words, by measuring the relative amounts of tRNAs it’s possible to tell whether the cell is dividing, as is the case with cancerous cells, embryonic stem cells and other stem cells, or whether it’s a healthy cell that has stopped dividing after having specialized into a particular tissue type, such as a bone, blood or skin. In fact, all cancerous cells in the study were found to have characteristic tRNA signatures, which could be further classified by cancer type.
Changes in behavior
When the scientists examined the connection between tRNAs and different types of genes, they found, unsurprisingly, that the tRNAs more often present in dividing cells tended to be involved in the translation of genes that govern more “individualistic” cellular behavior – for example, rapid growth that doesn’t take neighboring cells into consideration – a feature typical of cancer. On the other hand, tRNAs present in non-dividing cells were associated with genes responsible for more “group-oriented” behaviors, such as cellular adhesion and communication, typical of healthy tissue.
The scientists think the two tRNA signatures could have evolved as one of nature’s many ways of trying to block cancer. These signatures represent a mechanism that may add to cellular stability by keeping the two states, dividing and non-dividing, separate. When a normal non-dividing cell is on its way to becoming cancerous, it needs to alter its tRNA repertoire, a task that presents yet another hurdle it needs to overcome before it can start dividing uncontrollably: until this repertoire is changed, the translation of genes with cancerous potential may be less efficient.
Whatever their evolutionary significance, the analysis of tRNA signatures may in the future be developed into a tool for detecting or diagnosing cancer or for making a clinical prognosis of cancer development. An improved ability to distinguish between cancerous and healthy cells may also facilitate the design of therapies that will selectively target the tumor while sparing healthy tissue.
Prof. Yitzhak Pilpel's research is supported by the Sharon Zuckerman Laboratory for Research in Systems Biology; and the Braginsky Center for the Interface between Science and Humanities. Prof. Pilpel is the incumbent of the Ben May Professorial Chair.