Are you a journalist? Please sign up here for our press releases
Subscribe to our monthly newsletter:
“The membrane surrounding a cell is very thin,” says Dr. Sarel Fleishman. “You can think of it as a soap bubble: If you try to isolate it, it will pop.” Yet this thin membrane is what separates the inside of every living cell from the outside. A sort of barrier with gates, the membrane is full of proteins spanning its width that transfer nutrients and signals from one side to the other. Indeed 30% of the proteins in our bodies are membrane proteins, and many drugs, including anti-cancer drugs, work by targeting these proteins. Thus a better understanding of these proteins’ molecular and physical features may contribute to everything from biomedical research to the design of artificial proteins and new drugs.

In research that was recently described in eLife, Fleishman and research students Assaf Elazar and Jonathan Weinstein, together with Prof. Eitan Bibi and his student Ido Biran, all of the Weizmann Institute’s Biomolecular Sciences Department, created, for the first time, a new method for investigating the interactions between a protein and its membrane within a living cell. Going down to the level of the protein building blocks – amino acids – and the genes that encode them, they were able to show that to remain stable within the membrane, proteins have to stick to a basic plan.
The membrane is made up of two layers of double-sided molecules that create water-loving outer and inner surfaces and an oily, water-repelling core. This arrangement produces a negative electric charge on the inner surface facing the cell. The membrane-spanning proteins come into intimate contact with these various layers, as well as with other proteins, both inside the membrane and at either end.
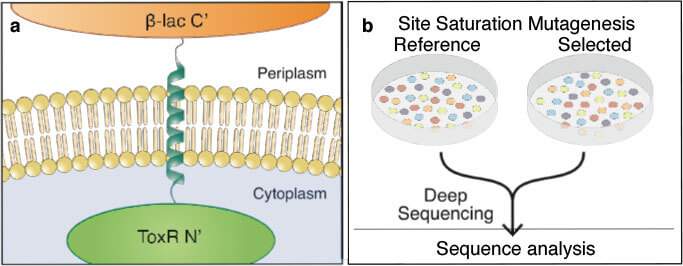
Fleishman and his group initially set out to understand how membrane-spanning proteins interact with one another within the membrane of a living cell. They combined experimental systems from two very different areas of the life sciences. In one, a certain protein is carefully mutated multiple times to generate hundreds of slightly different variants. Genes encoding these mutated proteins are fused to genes for antibiotic resistance and inserted into bacteria, which are then grown all together in a dish. When the bacteria are exposed to antibiotics, those mutants that have effective protein-membrane interactions outgrow those that do not. Afterward, the researchers employed a type of genomic sequencing called deep sequencing that enabled them to distinguish between the different mutants and count how many daughter cells of each survived the antibiotics – in other words, which mutants made more successful proteins.
We managed to provide real numbers to outline several of the basic rules for building membrane-spanning proteins
After plotting the relative amounts of hundreds of different mutations – for each of the 20 amino acids in each of around 20 different spots on the protein structure – the group realized that what they were looking at was essentially a map explaining which amino acid should go where in order for a protein to be stable within the membrane. Somewhat to their surprise, the detailed information about membrane-protein structure revealed in this map explained certain theories of membrane composition. So, for example, they identified the positively-charged amino acids that will generally find themselves the sections of the membrane that have a net negative charge – those facing the cell – as well as those neutral amino acids that will show up more toward the central, oily, segments. Although evidence for this basic idea had been found over several decades in a number of “qualitative” studies, this is the first time that such quantitative measurements had been produced, providing real numbers for the structure of proteins in the membranes of living cells.

“By bringing together ideas and methods from chemistry and biology,” says Fleishman, “we managed to provide real numbers to outline several of the basic rules for building membrane-spanning proteins. Our data have actually resolved several vexing dilemmas concerning the nature of biological membranes.” He and his group are already envisioning several directions in which this new insight may take them. Fleishman’s group focuses on creating artificial proteins through computer algorithms, and these new rules should help them design new protein structures for various functions. These rules may also help researchers design better drugs to target membrane-spanning receptors. Such drugs typically aim for either the outer regions of these receptors or the parts that are inside the cell, but; Fleishman and his group think that there may be opportunities to target the proteins within the cell membranes, as well.
Dr. Sarel-Jacob Fleishman's research is supported by the Yeda-Sela Center for Basic Research; the Rothschild Caesarea Foundation; Sam Switzer, Canada; and the European Research Council. Dr. Fleishman is the incumbent of the Martha S. Sagon Career Development Chair.