Are you a journalist? Please sign up here for our press releases
Subscribe to our monthly newsletter:
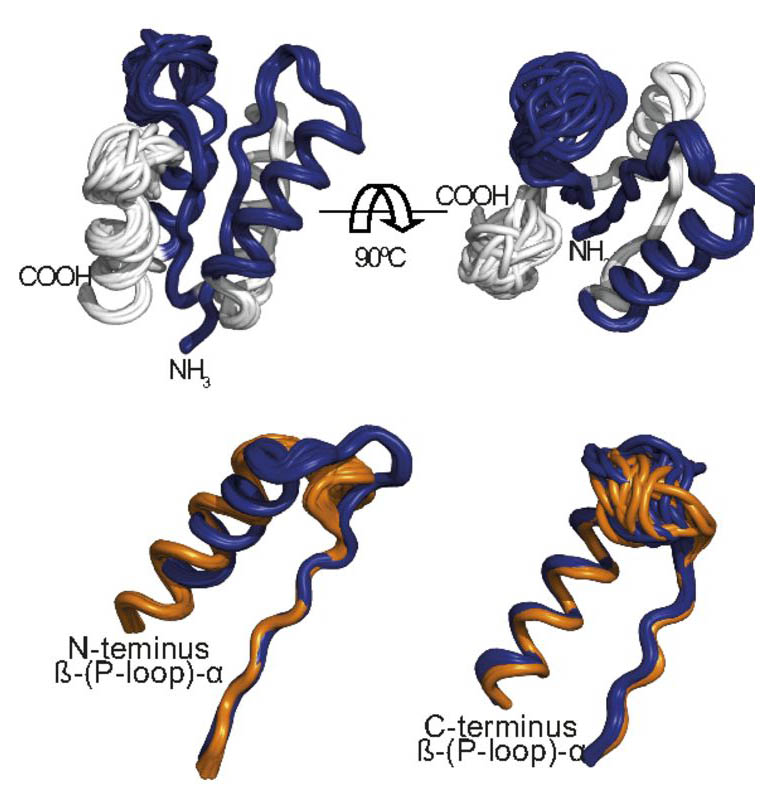
Even the lowliest bacterium houses a complement of sophisticated enzymes – proteins that carry out the basic tasks of life. We know that enzyme complexity, with numerous active sites working together to complete specific tasks, has ancient origins. How did this complexity arise? This question has remained open, in part because any traces of primordial enzymes have long been erased from the historic record, and in part because it has been hard to envision how a “simple” enzyme – one with few working components – might function. Now scientists at the Weizmann Institute of Science and their colleagues have provided a possible answer with a protein they created in the lab. Their experimental findings may shed new light on the very early stages of cellular life, some 3.7 billion years ago.
The study, which was recently reported in the Proceedings of the National Academy of Sciences (PNAS), was led by Dr. Maria Luisa Romero-Romero in the group of Prof. Dan Tawfik of the Institute’s Biomolecular Sciences Department. The group included the lab team of departmental colleague Prof. Michal Sharon, as well as those of Profs. Gabriele Varani and David Baker of the University of Washington and researchers from Germany, Singapore and Norway. The scientists based their initial idea on existing studies and years of experience that suggested the place to begin creating a simple enzyme would be the so-called phosphate-binding loops, or P-loops, found on many different enzymes, in everything from bacteria to humans. These short loops, around eight amino acids in length, work directly with another molecule found in all living things: ATP.

ATP is the main currency of the cell, and it has been around even longer than proteins. It is part of the cell’s basic energy cycle, recycling energy and driving motor proteins, pumps and muscle fibers. Enzymes pluck phosphates off the ATP molecule to use in the various biochemical reactions they implement. These reactions mainly involve attaching the phosphate groups to other metabolites like sugars, activating them like coin-op machines. So the P-loop, which carries and releases the phosphate group, is crucial to the function of many different enzymes.
That is why the researchers hypothesized that a prototype of an early enzyme could be fashioned out of a P-loop, even though experiments had shown it does not normally function independently. They began by using the sequences of modern P-loop-based proteins to infer the oldest P-loop sequence, and they then designed brand new protein sequences to contain this ancient loop. The criteria for these structures: To be as short as possible and to be made up of an “elementary” minimal set of amino acids while still enabling the P-loop to function, and to have no phosphate binding activity themselves.
These sequences were tested in lab dishes. Eventually the researchers found several P-loop-bearing sequences that were active with only 55-100 amino acids. That is something between a third and a sixth of the length of an average enzyme today. More importantly, the only functional element on the entire enzyme was the P-loop. “The fact that these small proteins worked so well helps us understand how enzymes could have started from short sequences, and these at some point started linking together and adding functions as life evolved,” says Tawfik.
Complexity – even when it seems to be a sort of irreducible ‘clockwork’– really evolves from simpler parts
The surprise, he adds, was that the P-loops in the designed proto-enzymes not only bound ATP but bound very strongly to DNA and RNA, something that the modern P-loop proteins do not do. In other words, their enzymes were taking on functions not seen in today’s enzymes.
“This may add a twist to a scenario many researchers are coming to accept in which the first life consisted of DNA and RNA, with the RNA performing the functions that enzymes carry out today,” says Tawfik. “These findings suggest that the first enzyme-like proteins may have been ‘assistants’ to the RNA and DNA. These would have been all-purpose and relatively crude, so as evolution progressed, they were replaced by enzymes that were more and more efficient and specialized.
“In essence, we showed that complexity – even when it seems to be a sort of irreducible ‘clockwork’– really evolves from simpler parts. It is as though an alien came to Earth and tried, by taking apart a car, to understand how this complex machine came to be. Our proteins, like cars, appear to us ready-made, and we don’t see the ‘carts’ and ‘wheelbarrows’ that preceded them. We think we have now demonstrated the enzyme equivalent of a wheelbarrow. As Darwin put it: ‘From so simple a beginning endless forms most beautiful and most wonderful have been, and are being, evolved’.”
If the above research went in search of proteins’ past, other recent research published in PNAS in which Tawfik and his team participated may well represent the future of proteins. A research team including Prof. Arren Bar-Even of the Max Planck Institute of Molecular Plant Physiology in Potsdam-Golm, Germany, and Dr. Sarel Fleishman of the Weizmann Institute’s Biomolecular Sciences Department managed to create a new enzyme pathway – one that might help plants use carbon dioxide (CO2) more efficiently. If plants can be engineered to produce this pathway, it might lead to higher crop yields in the future.
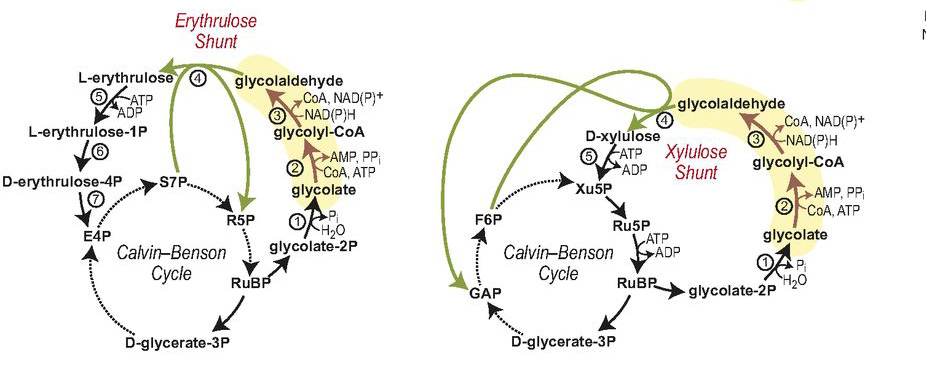
Plants, as we know, take in CO2 from the atmosphere and convert it, with the aid of photosynthesis, to carbon-based proteins and sugars (carbon fixation). This not only provides all other life on the planet with food and free oxygen to breathe, it keeps the levels of CO2 naturally low.
The part of the cycle the researchers decided to improve upon is a sort of sideshow to the main event of carbon fixation – specifically, to its central enzyme, Rubisco. During photosynthesis oxygen is released; but Rubiso can bind to the free oxygen in a side reaction that is harmful to the plant. To prevent this from happening, a secondary enzymatic pathway degrades the oxygen product of Rubisco, a compound known as glycolate. This process uses quite a bit of the plant’s energy and can be wasteful of CO2, even releasing it back into the atmosphere. The researchers decided to design a new enzyme pathway that would recycle glycolate: Rather than being broken down, glycolate is reused and its carbon goes into sugars rather than being released into the atmosphere as CO2.
This pathway, because it does not exist in nature, was designed from beginning to end. Ultimately, the team produced two new enzymes that work together with two natural enzymes in recycling glycolate into reusable parts (in contrast to the natural plant glycolate degradation pathway, which involves over 10 enzymes). This pathway worked in the lab dish just as computer models had predicted, in a process that was efficient – using roughly half of the energy plants use to get rid of glycolate – and keeping the CO2 release to a bare minimum. The researchers hope to eventually insert the entire glycolate-recycling pathway into photosynthetic bacteria or plants. Among others, the project is supported by a European Union’s Horizon 2020 FET Programme Grant, with the intent of helping the world increase crop yields and fight anticipated shortages in the food supply.
Prof. Dan Tawfik's research is supported by the estate of Bill Danczinger. Prof. Tawfik is the incumbent of the Nella and Leon Benoziyo Professorial Chair.