Doctors seeking to prescribe personalized therapy for a cancer patient face a great deal of uncertainty. How fast is the tumor growing? How aggressively should the cancer be treated? And which treatment approach is likely to be most effective?
Weizmann Institute scientists propose an original and relatively simple method for making sense of the vast amounts of cancer-related genomic data.
Prof. Eytan Domany and Dr. Yotam Drier of the Physics of Complex Systems Department have developed
an algorithm called Pathifier that can help medical researchers and practicing physicians assess the prognosis of a given tumor based on its genetic profile. As reported in the
Proceedings of the National Academy of Sciences (PNAS), the scientists, together with postdoctoral fellow Dr. Michal Sheffer, have already used Pathifier to identify previously unknown cancer subtypes that differ in their prognosis.
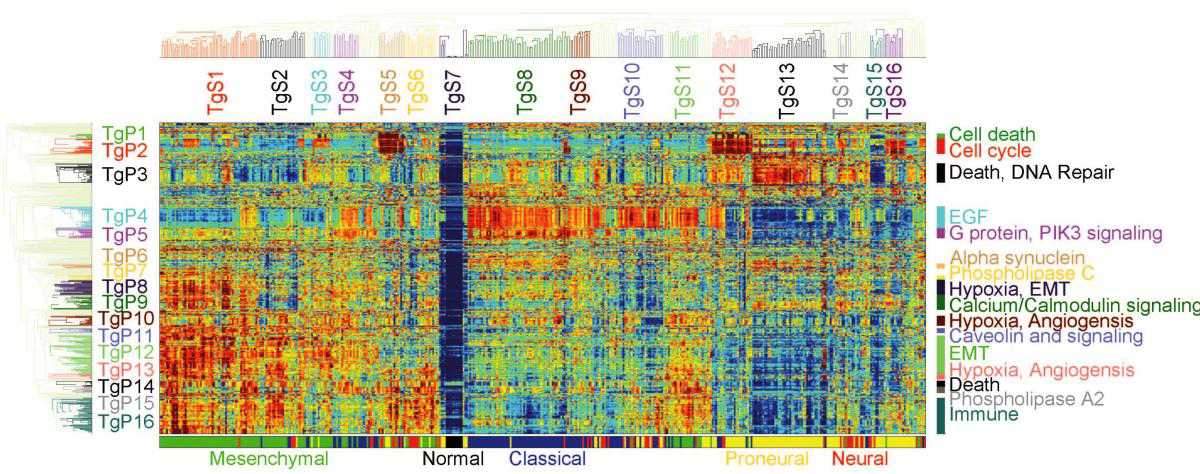
Pathifier, as its name suggests, analyzes pathways, the biochemical processes making up the life of the cell, each involving 20 to 30 genes. About 500 such pathways are known to science. In any given cancer, at least a few of them are abnormal. For example, if some of the pathways that govern growth are defective, the cell keeps dividing uncontrollably, leading to the formation and growth of a tumor. The extent of deregulation of different pathways varies from cancer to cancer and from patient to patient.
By comparing genomic data from cancerous and healthy cells, Pathifier assigns a “deregulation score” to each pathway. The set of these scores makes up the profile of the tumor. The researchers believe it can help evaluate the cancer’s aggressiveness, assess its chances of responding to a particular therapy and perhaps even identify key biochemical processes that in the future may serve as targets for therapy.
Because the new method focuses on entire pathways, it gives a more accurate picture of the cancer’s properties, as opposed to monitoring individual genes. “It’s like analyzing what’s wrong with a car by observing the performance of its engine, brakes, steering and other systems, rather than dismantling it and looking at all the individual nuts and bolts,” Domany says. Moreover, the method is particularly reliable because it is based on the analysis of large sets of genomic data from hundreds of patients. Yet it is manageable because it focuses only on essential data, rather than trying to encompass the entirety of genomic details.
In the new study, the researchers applied Pathifier to a malignant brain tumor called glioblastoma. It was known from past genetic analyses that patients with a certain type of this tumor survive longer than others, but on the basis of the tumor profiles, the algorithm has allowed scientists to identify a smaller subtype of patients who are the truly longer survivors, as opposed to the rest of the patients with the same tumor type, who are not. Furthermore, Pathifier has enabled the scientists to identify three pathways whose levels of deregulation are strongly indicative of the survival prospects of colon cancer patients.
Currently, Pathifier can be used as a research tool, offering medical researchers a reliable way of processing cancer-related genomic data. In the future, the algorithm can point to relevant biomarkers – that is, measurements of the levels of certain chemicals that are indicative of the activation of key pathways – that could help practicing physicians choose appropriate treatments for their patients. The physicians could rely on such biomarkers to evaluate a tumor’s deregulation profile.
Prof. Eytan Domany's research is supported by the Kahn Family Research Center for Systems Biology of the Human Cell, which he heads; the Mario Negri Institute for Pharmacological Research -Weizmann Institute of Science Exchange Program, the Leir Charitable Foundations; Mordechai Segal, Israel; and the
Louis and Fannie Tolz Collaborative Research Project. Prof. Domany is the incumbent of the Henry J. Leir Professorial Chair.