- Putting polymers to practical use
- The mechanochemical machine
- The physics of biology
- Supermodels: Useful as well as elegant
- Enzyme bonding that's become important in drug and food production
- When "V" means "Very good fit"
- Digesting the indigestible
- The molecular mechanism of high blood pressure
- How to catch a molecule
- Spots that hate water
- Computerized molecular recognition
- Hard work at the protein factory
- Chemical brakes for biological processes
- 3-D protein images that help fight disease
- The strongest biological glue
- How to live in the Dead Sea
- One smell after another
- The energy that powers living things
- Down to the bone
Putting polymers to practical use
Throughout the Institute's early decades, Weizmann scientists investigated the structure, properties, and functioning of polymers-long molecules built out of strings of smaller subunits. DNA, proteins, polysaccharides and synthetic plastic materials are all polymers. Among other things, the scientists studied the properties of electrically-charged polymers (polyelectrolytes) and their interactions with ions and other small molecules. This research led to the development of ways of using polymers in the food and pharmaceuticals industries, as well as improving soil properties for growing a variety of plants for agriculture.
The mechanochemical machine
In the 1950s, an Institute scientist managed to build a mechanochemical machine involving fibers of the protein collagen which is found in the bone, tendons, and skin of all animals. When subjected to different dissolved salts, the fibers expanded or contracted, thereby operating a set of wheels. This piece of research significantly advanced scientists' ability to investigate and understand the physical background of fundamental life processes.
The physics of biology
Institute scientists were among the first to apply their knowledge of physics to research into biological systems. A key contribution was in advancing the understanding of how the principles of irreversible thermodynamics are expressed in biological systems. These are processes from which there is no return, such as fetal development, aging and death. On the basis of these studies, carried out in the 1950s, subsequent research was able to provide an understanding of vital life processes such as the transport of ions across the membranes of cells.
Supermodels: Useful as well as elegant
Proteins have a highly complex structure which includes long, twisted chains containing sequences of up to 20 different amino acids. To make it easier to study the properties of proteins, an Institute scientist in the 1950s developed simple synthetic models, known as polyamino acids, containing just one or two different amino acids. These models, which continued to be used throughout the 60s and 70s, were an important tool in determining the spatial structure of protein molecules using X-rays.
The synthetic models were also used for research into the processes of degradation, destruction, and disintegration of the spatial structure of proteins. They were also of use directly and indirectly in the cracking of the genetic code.
This understanding of the properties of polyamino acids, and above all of their immunological properties, enabled the development of synthetic antigens, which, among other things, led Weizmann scientists to develop the drug Copaxone (Copolymer 1), now used worldwide to treat people suffering from multiple sclerosis. Copaxone is a relatively simple synthetic copolymer, made up of four amino acids, which are the building blocks of proteins.
Enzyme bonding that's become important in drug and food production
In the 1960s, a Weizmann Institute scientist developed a method of adsorbing (bonding) as well as covalent binding of enzymes to a variety of insoluble carriers. Enzymes catalyze (speed up) many different chemical processes and adsorption or binding enables them to be reused time and again, without having to go through the lengthy and tricky process of separating them from the materials on which they act. Furthermore, as the technique also stabilizes enzymes, immobilized enzymes have become an important tool in the pharmaceuticals industry. Among other things, they are used to manufacture penicillin and other antibiotics. Another application is in separating chiral pharmaceuticals - molecules with identical chemical composition but with two possible structures that are "mirror images" of each other. Often, one form of the molecule is effective against a disease while the other is not.
The immobilized enzyme techniques also form the basis of various processes in the food industry, for example, in the conversion of starch into a fructose-enriched syrup which is widely used in the production of candies and soft drinks.
When "V" means "Very good fit"
Weizmann Institute scientists discovered that molecules of various sugars and specific sugar-binding proteins (called lectins) present on living cells function like a "V" connection, allowing the cells to identify one another and latch on. This type of recognition is responsible for joining sperm to eggs as a prelude to fertilization, for binding bacteria to host cells in the first step of infection and white blood cells to the interior wall of blood vessels, which is an important step in fighting infection and inflammatory processes.
The "V" connection is similar to the way a key fits into a lock and is so sensitive and precise that it can even distinguish between two molecules of highly similar sugars, such as glucose and galactose.
Weizmann researchers demonstrated that lectins are found not only in plants and animals, but in bacteria too, and that the lectin-sugar match plays, as mentioned, a crucial role in the binding process of disease-causing bacteria to human tissue. Also, nomadic cancer cells on their way to establishing new metastases (growths) in the body carry an "anchor" in the form of a lectin that binds galactose. This anchor allows them to "land" in new sites in the body and create metastases.
These contributions by Institute scientists to understanding the roles of sugars in the external relations of living cells and to elucidating the precise structure of the lectins and the way in which they bind sugars, have paved the way to developing treatments for various diseases. These involve using alternative, decoy sugars to bind to the lectins, thereby blocking or regulating their ability to bind to sugars on the cell surface.
Weizmann scientists, in collaboration with researchers at Tel Aviv University, demonstrated for the first time in experiments with mice that these decoy techniques can be used to destroy the ability of disease-causing bacteria to bind to body tissues (which is the first stage of infectious disease). Several pharmaceutical firms are developing applications based on this approach for the prevention and treatment of bacterial infections in humans.
In red blood cells, sugars on the cell surface determine the different blood types such as A, B, AB, and O. Institute scientists proved that removal of one sugar molecule from type B red blood cells actually converts them into type O, suitable for transfusion to people with any blood type. This could help overcome temporary shortages of specific blood types in emergency situations.
The binding properties of sugars and lectins are useful for the characterization and separation of different types of white blood cells, as well as bone marrow cells. This is extremely helpful in transplants. By binding specific lectins to cell sugars, it is possible to identify and then remove the damaging white blood cells from donor bone marrow. This is important because damaging white blood cells in donor bone marrow may prevent the body from accepting the bone marrow transplant. Today, this lectin method for bone marrow purging is routinely used for saving the lives of children born with immune deficiencies, also known as "bubble children," and on an experimental basis for adult leukemia sufferers.
Digesting the indigestible
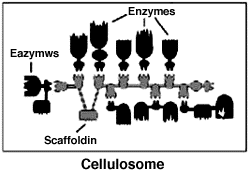
Weizmann Institute researchers and their colleagues discovered a multifunctional, extracellular organelle that they have called the "cellulosome." They have recently uncovered important structural features of the cellulosome, including how it binds to the cellulose substrate and how the various subunits are incorporated into the multicomponent complex.
The cellulosome is responsible for the efficient breakdown of cellulose into simple sugars (disaccharides). This decomposition is the first stage in the process many animals use to digest and produce energy from the plants they eat. The discovery may lead to diverse applications, including the exploitation and management of cellulose waste, controlled-release drug systems, diagnostic tests for various materials, genetic tests and more.
The human body breaks down complex sugars (polysaccharides) like starch, and disaccharides like lactose and sucrose into simple sugar units of glucose, which is the main source of human energy. However, the human body cannot digest cellulose, which is a polysaccharide with a much more stable structure. This means that humans cannot exploit the glucose it contains, and cellulose passes through the body as dietary fiber. In contrast, ruminant animals, such as cows, with the aid of bacteria found in their digestive tracts, can degrade cellulose and use it as an energy source.
When breaking down cellulose substrates, many bacteria use their cellulosomes - structures composed of numerous, different proteins (many of which are enzymes). Weizmann Institute scientists discovered that the key to efficient cellulosome activity is a scaffold-like protein called "scaffoldin" which mediates between cellulose and the enzymes which act on it. They are currently attempting to clone various combinations of the components of scaffoldin (via genetic engineering) and have already produced one combined enzyme system that is extremely effective in decomposing cellulose and other cellulose-containing substances.
The ability to produce scaffoldin structures that can form links between different substances and enzymes may allow controlled implementation of many and varied biochemical processes. This should create the basis for an entirely new field of biotechnology.
The molecular mechanism of high blood pressure
Hypertension (high blood pressure) stems from a combination of environmental and hereditary factors and occurs in about 15 percent of the adult population in developed countries. It is a major cause of heart disease, kidney problems and stroke.
One major cause of hypertension is a malfunction in the natural control mechanism which enables the kidneys to filter the blood and regulate the body's concentration of salts and water. Increased absorption of sodium salts leads to excess absorption of water (as the body tries to dilute the salts) which heightens the pressure that blood exerts on the walls of the arteries.
Weizmann scientists have contributed to the understanding of the natural processes that regulate sodium salts in the body by demonstrating the central role of two proteins: the "sodium channel" and the "sodium-potassium pump" (Na/K pump or Na/K-ATPase). The researchers followed up these studies by indicating methods of regulating blood pressure more effectively.
Sodium channels are molecules located in the cell membranes of the kidneys, where they transfer sodium ions into or out of the cells. The researchers found that the channels are regulated by a steroid hormone secreted by the adrenal gland. This hormone penetrates the kidney cells where it binds with a specific receptor, thereby prompting two processes: the opening of new channels which previously were not active, and an increased formation of new channels.
Determination of the structure and function of the sodium-potassium pump by the Weizmann Institute scientists is helping to better understand the way the protein interacts with steroid-like cardiac glycosides, implicated in the generation of hypertension. Additional studies have found that a hereditary tendency to develop high blood pressure may be due to a genetic defect that leads to the overproduction of sodium-potassium pumps.
How to catch a molecule
Since the 1970s, affinity chromatography has served as an important tool in all biotechnological fields, including genetic engineering, and advanced fields of medicine and biology. The technique - which is a method for separating biological materials such as enzymes, antibodies, hormones and receptors - was developed by Weizmann Institute scientists.
Many biological studies require that different molecules be separated (either for identification, or for use in further reactions or processes) from a compound mixture consisting of tens and even hundreds of types of molecules. Until 1968, many years of research were often needed to isolate a few milligrams of a substance. Weizmann Institute researchers resolved this through their understanding that many proteins have a natural partner that binds specifically to them. When such a partner does not exist naturally, it is possible to prepare it artificially or to create an antibody that acts on the protein.
The Weizmann technique involves isolating a sufficient amount of one of the pair, then binding it to a solid matrix and using it as "bait" for its mate. The sought-after substance binds to its partner and is "released" for use (if required) by chemical techniques.
Spots that hate water
Weizmann Institute scientists developed a new methodology called hydrophobic chromatography to characterize, separate and isolate proteins and cells. This method is based on hydrophobic (water-repelling or water-hating) hydrocarbon chains of different lengths and shapes, which are anchored on inert beads and interact with hydrophobic "pockets," "patches" or "spots" found on the surface of various proteins. Such hydrocarbon chains interact with protein molecules submerged in water and bind them to minimize the surface of the hydrophobic area that is in contact with the water.
It turns out that proteins can be distinguished and resolved from one another since they differ in the size, number, and water repellence of their hydrophobic patches or pockets, i.e., since every protein has its own unique set of hydrophobic spots.
Weizmann Institute researchers developed a series of such beads that allow the separation and identification of proteins. This technique (under different names) turned out to be a very powerful method of separation and has been used throughout the world for research in biochemistry, biology and medicine, as well as in the biotechnology industry. It is used in the preparation and characterization of drugs such as enzymes, hormones, antibodies and receptors.
Computerized molecular recognition
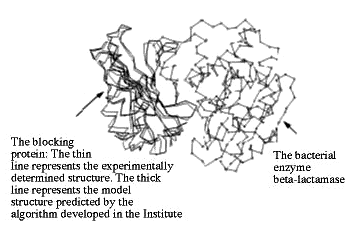
A multidisciplined Weizmann Institute research team developed a novel algorithm for forming three-dimensional computer models of various molecules, comparing one with the other and examining the ways they can recognize or "dock" with each other. Because molecular recognition is essential in all life-sustaining processes, understanding it may advance new and exciting insights into biochemical processes and encourage the development of new drugs.
The algorithm developed by the Weizmann scientists has already successfully predicted how a specific protein identifies, binds with and blocks an enzyme that makes bacteria resistant to antibiotics.
Hard work at the protein factory
Weizmann Institute researchers, in cooperation with colleagues at the Max Planck Institute, were the first to crystallize ribosomes, the organelles that are responsible for protein synthesis in all cells of microorganisms, animals and plants. Immensely powerful X-rays are now being used to decipher the structure of the ribosomes and provide clues as to how they work.
An analysis of parallel imaging studies has already produced several important revelations, e.g., that ribosomes contain a "tunnel" that appears to protect newly-formed proteins. They also identified the groove along which travels the mRNA, the messenger molecule that carries the genetic information of the DNA about the proteins to be synthesized. They identified the space where the biochemical activity connected with protein synthesis takes place.
Chemical brakes for biological processes
Weizmann Institute scientists have contributed to understanding the activity of enzymes, the large protein molecules responsible for many chemical reactions that are vital to life. An enzyme functions via its active site - the part of the enzyme that comes into contact with the molecule or molecules on which it works. This is known as the substrate.
Institute scientists demonstrated that the active site of proteolytic enzymes is much larger than was commonly thought. The scientists then developed a model and method to map out the active site by measuring its size and defining its chemical properties. This mapping allows for the rational design and synthesis of substances, known as inhibitors, which specifically retard enzyme activity. This is an important means for enzyme research: It helps to understand how different molecules recognize each other, and also allows for the development of medications based on inhibiting a variety of proteolytic enzymes involved in the pathogenesis of many diseases, such as AIDS.
3-D protein images that help fight disease
Weizmann Institute scientists have significantly improved the usefulness of the world-wide databank of protein structures that is available on the Internet. In cooperation with colleagues at the Brookhaven National Laboratory in the United States, the effective use of this databank has been made possible. The databank carries detailed information on the 3-D structure of more than 7,000 proteins and nucleic acids. The 3-D structure of a protein is a critical factor in its activity.
Therefore, making this information available around the world should help researchers working in the growing field of protein regulation, e.g., in drug design.
Proteins are key functional elements of living organisms, including, of course, human beings. An excess or shortage of a given protein or a small genetic change in a protein may cause physical or mental problems, disease, or even death. For these reasons doctors, scientists, and drug companies are devoting a major effort to working out the 3-D structures of proteins and to understanding how they work and how they are regulated.
The ability to control a specific protein depends largely on understanding its spatial, or 3-D, structure. Once the exact structure is known, a suitable drug can be tailored to bind to the protein and neutralize its activity. However, proteins have a very complex structure, consisting of a long and twisting chain of different amino acids. Ascertaining this structure is a lengthy, complicated and very expensive business, aided in recent years by X-ray synchrotron radiation and nuclear magnetic resonance (NMR) imaging.
Researchers developing methods of regulating the activity of proteins need specific information about their structure; this data has often been developed by scientists working in distant locations. Until recently, such information had been stored in particularly powerful and fast computers and could not be transferred via the Internet using standard communication lines. Researchers had to make do with text or numerical lists only.
Now, Weizmann and Brookhaven have developed a computerized system that bridges these limitations by transferring precise structural information via communication lines to the Internet. This enables a researcher to retrieve a 3-D image of a protein molecule quickly, even within seconds, on his or her personal computer.
The strongest biological glue
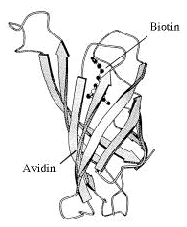
Weizmann Institute scientists discovered the strongest biological "glue" known to nature, based on the structural and chemical compatibility between the protein avidin (found in eggs) and the vitamin biotin. This property has been used for years to bind biological materials together in many diverse research procedures.
However, the strong bond between avidin and biotin created difficulties for researchers trying to break this adhesion, and, to a certain extent, caused waste. Therefore, the Institute's scientists developed a new method to break the link, by changing one of the many hydrogen bonds which cause the avidin molecule to recognize the biotin. This discovery significantly enhanced the efficacy of one of the most important research tools used in life science studies worldwide.
How to live in the Dead Sea
Weizmann Institute researchers have uncovered one of the mechanisms which may help a particular species of bacteria to survive in the salt-rich environment of the Dead Sea, where most organisms would ordinarily die. This was done by comparing the properties of a particular protein in that bacteria with the properties of its counterpart in freshwater bacteria.
The investigators grew crystals of the protein ferredoxin from the Haloarcula marismortui bacterium, which flourishes in the Dead Sea. The crystal was then irradiated with X-rays to find its exact structure. This revealed that the ferredoxin from Dead Sea bacteria has an additional area that does not exist in ferredoxin of non-saltloving bacteria. This area and the rest of the surface of the molecule have a relatively strong negative electrical charge and therefore attract water molecules which surround the protein, forming a dense protective layer. This protective envelope appears to be connected with the bacteria's ability to live in the harsh environment of the Dead Sea.
Understanding how life survives in uninviting surroundings such as the Dead Sea could help scientists to modify organisms, thus enabling them to survive in difficult environments that might eventuate on the Earth now and in the future.
One smell after another
Weizmann Institute scientists discovered an enzyme-controlled mechanism that is apparently responsible for weakening and removing individual smell sensations, a vital process that enables our noses to continually sense new odors. This "shutdown" mechanism proposed by Institute researchers involves two enzymes that work on odor molecules one after the other. In the first stage, the enzyme cytochrome P-450 oxidizes the smelly material by the addition of a hydroxyl group. This activates it, facilitating the occurrence of a variety of subsequent reactions.
At a later stage the second enzyme, glucoronyl transferase, links the odor material (which is usually a water-avoiding hydrophobic molecule) to a molecule of glucoronite acid (a water-loving hydrophilic molecule). This bonding alters the odor molecule, creating an incompatibility between it and the protein receptor to which it was previously bound. This incompatibility weakens the link between the receptor and the odor material, and so we stop smelling it; it also facilitates clearance of the odor in the blood stream.
The energy that powers living things
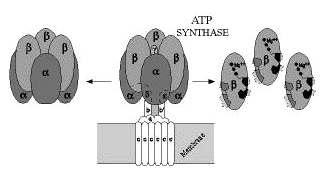
Weizmann Institute scientists isolated a subcomplex of the enzyme ATP synthase which forms ATP (adenosine triphosphate), the principle energy-carrying molecule in all biological systems. ATP is a sort of common currency by which it is possible to exchange and convert energy into different forms according to the needs of every living cell. A human body at rest requires about 40 kilograms of ATP a day, but at any one time contains only a minute amount of it, since it turns over very fast.
Knowledge of the enzyme's structure and how it produces ATP should open new and broad fields of medical and industrial applications. However, ATP synthase, which is the largest enzyme identified to date, is in no hurry to reveal its secrets. It is known to consist of two main parts: one part serves somewhat like a pedestal, permanently set in the membrane of the mitochondrion (in animals) or of the chloroplast (in plants); the second, attached to the pedestal, protrudes into the cytoplasm which fills the internal space of every cell. This is the active part of the enzyme and is built from five different-sized subunits. There are three copies of each of the two large subunits, arranged alternately around a central cavity, like an orange with six segments. There is one copy of each of the three small subunits; the largest is located inside the central cavity and is linked to the other two, binding the "orange" to the pedestal.
From a chloroplast membrane, Institute researchers isolated the orange with its six segments, which contain three catalytic sites where ATP is produced. Later, the scientists isolated two genes, each encoding one of the two subunits forming the orange's segments. Then, using genetic engineering techniques, they produced large quantities of these two subunits in bacteria. Today, this biotechnological production system is being used to identify by site-directed mutagenesis the amino acids that are essential for assembly of the orange and formation of the catalytic sites where ATP is synthesized. Recently an ATP synthase, having only one different amino acid in one of the two subunits forming the orange segments, was found to produce ATP in larger quantities than does the natural enzyme.
Down to the bone
Weizmann Institute scientists have made an important and unique contribution to the understanding of the internal hierarchical and complex composition of bone structure.
The researchers examined a common type of mammalian bone that is characterized by an internal structure based on a group of onion-like multilayered cylinders, with a blood capillary passing through its center. Focusing on the internal structure of the microscopic "onion skins," they discovered that the layers of the protein collagen and the calcium phosphate crystals in the spaces between them organize themselves like a natural composite material.
A composite material is made from two substances, one of which serves as a sort of mold that is filled with the other substance. Current uses of synthetic composite materials include airplane wings, satellite parts, bicycle helmets and even kitchen utensils and innovative tools.
In bone, the collagen fibers create a mold or matrix in which calcium phosphate forms. Thus, a multilayered structure reminiscent of plywood is formed. The scientists discovered that the layers of fibers are laid one on top of the other at angles that vary in every layer at a fixed rate of about 30 degrees. This general structure repeats itself every five layers.
These findings led to the development of a mathematical model that enabled researchers to better elucidate the connection between the structure of bone and its unique mechanical properties. It is hoped that these studies will lead to a better understanding of normal bone function, and, in turn, the many serious problems that occur when bone malfunctions.